Edinburgh/DivisionPopper/References
From 2007.igem.org
MENU : Introduction | Background | Applications | Design | Realization | Modelling | Status | Conclusions
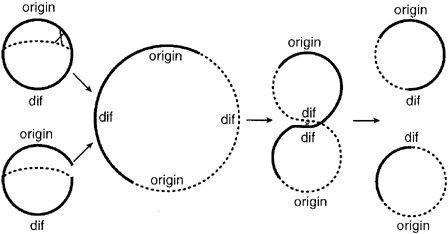
Some of the biological mechanisms at the base of the Division PoPper design have been widely studied in the past and well reported in literature, as for example the repression mechanism of promoters Teta and CLambda. Some others are instead quite new and not yet completely understood. In particular this is true for the dif recombinase system and its relation with the cell division. Our assumption that dif sites can be used as physically mechanism to "flip" a DNA region during cell division found positive indication in some published papers. With this project, and in particular with the proof of concept system realization and analysis, we hope to gain a better understanding of the relation between cell division and recombinase mechanisms in order to plant the basis for further investigations. In this section we propose a brief literature review for dif recombinase system and cell division.
Contents |
[http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1449051&blobtype=pdf Bloor]
- An Efficient Method of Selectable Marker Gene Excision by Xer Recombination for Gene Replacement in Bacterial Chromosomes (2006)
- This article describes a system where a gene is deleted by replacing it with an antibiotic marker. The antibiotic marker is then excised during next cell divsion using the naturally expressed XerCD recombinases. This is similar to what we are doing except instead of excising we are inverting.
| "The natural dif site is in the chromosome terminus region and is essential for correct chromosome segregation at cell division: deletion of difE. coli results in a subpopulation of E. coli cells with a filamentous morphology (12). However, if natural difE. coli is deleted, another difE. coli site will not enable dimer resolution if placed outside of the 30-kb dif activity zone (DAZ) in the terminus region (6, 16). Therefore, while our method involves insertion of one or more extra dif sites at different chromosomal loci, this should not have an adverse effect on chromosome segregation, although the possibility that introduction of multiple dif sites in close proximity might result in deletion of intervening chromosomal DNA should be considered." |
- Summary: Dif sites outside of the DAZ zone shouldn't cause problems with the natural dif sites of the chromosome/plasmid
[http://www.blackwell-synergy.com/doi/full/10.1046/j.1365-2958.1998.00651.x Kuempel]
- Cell division is required for resolution of dimer chromosomes at the dif locus of Escherichia coli (1998)
- Good overview of the natural Xer recombinase system.
| "In strains deleted for dif, failure to resolve dimer chromosomes leads to the Dif phenotype, which includes filamentation in a fraction of the cells, abnormal nucleoid morphology, SOS induction and decreased growth rate and plating efficiency compared with wild-type cells (Kuempel et al., 1991; Cornet et al., 1996)." |
- Summary: Missing or broken dif sites result in filamentation and slower growth rate
| "dif is composed of a 28 bp ‘core’ sequence homologous to cer, which acts as the binding site for the resolvase enzymes XerC and XerD (Blakely et al., 1993). A difference between the sites is that cer is flanked by accessory sequences, which bind additional proteins and enhance recombination between sites in dimers. This provides directionality so that resolution of dimers to monomers is highly favoured (Blakely and Sherratt, 1996). Flanking sequences are not involved in resolution at dif as the phenotype of a 173 kb deletion can be suppressed by insertion of a 33 bp dif sequence (Tecklenburg et al., 1995). It has been proposed that directionality is provided by some aspect of chromosome partitioning itself. Structure is also important, as ectopic dif sites are only fully functional when located in an ≈30 kb region that surrounds the normal location of dif (Cornet et al., 1996; Kuempel et al., 1996)." |
- Summary: cer sites use accessory proteins to enhance dimer->monomer versus the reverse. dif sites doen't use accessory proteins? (what about FtsK?) and favor the dimer-> monomer direction by some other structural or mechanical mechanism inherent in cell division.
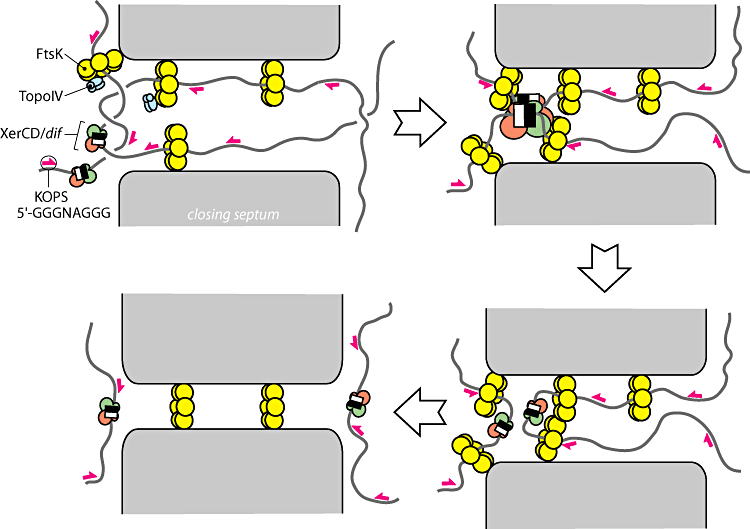
[http://www.blackwell-synergy.com/doi/pdf/10.1111/j.1365-2958.2007.05755.x Bigot]
- a literal chromosome segregation machine (2007)
- An up to date review on the mechanics of FtsK
- Summary: The yellow rings are the FtsK motors which powerfully pull the DNA through them. When they encounter dif sites with XerC and XerD recombinases on them the recombination event is activated