Wisconsin/Project
From 2007.igem.org
Contents |
Project Overview
Reprogramming gene expression is one of the main goals of synthetic biology. In the past, most groups have focused on cis-regulatory elements such as promoter to control transcription. We decided to explore artificial transcription factor (ATF), a trans-regulatory element, for controlling genes. Our project consists of characterizing ATF and testing ATF in mammalian cells.
Artificial Transcription Factor
Background
Artificial transcription factor (ATF) has two domains: DNA-binding domain and regulatory domain. The DNA-binding domain provides sequence specific targeting and the regulatory domain can up- or down-regulate gene expression. ATF design is modular, meaning one can mix and match different DNA-binding motifs with various transcription factors.
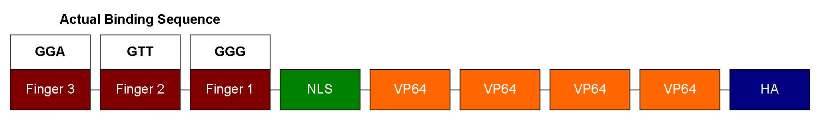
We decided to use zinc fingers for DNA-binding. A zinc finger is made of about 30 amino acids and is stabilized by a single zinc ion to form a certain protein structure (eg. beta-beta-alpha). It can interact with a specific 3 base-pair motif in DNA (Segal & Barbas, 2001). Regulation of gene activity is achieved by using zinc finger to recognize a specific DNA sequences, bind to the DNA, and recruite other molecular machinery that will activate or repress the gene. Our ATF project has centered on characterizing DNA binding specificity and cooperativity effects.
Design
Our ATF design has 3 zinc fingers followed NLS and VP64. NLS is for localizing ATF to the nucleus and VP64 is a strong transcription activator. We also performed site-directed mutagenesis to change the zinc finger recognition sequence.
Cognate Site Identity (CSI) Microarray
We are using CSI microarrays to quantify how well zinc fingers bind to DNA sequences. The CSI array contains every permutation of 9bp DNA sequence. This allows us to test the entire sequence space in an unbiased manner.
BCL-2 and Cell Fate Regulation
Background
Doxrubicin is a widely used drug to treat cancer. However one main limitation of its use is cardiotoxicity. Accumulated dose of doxrubicin can cause oxidative stress to cardiomyocytes and trigger apoptosis. Our goal is to up-regulate BCL-2 (a pro-survival protein) and prevent cells from dying under mild stress.
Design
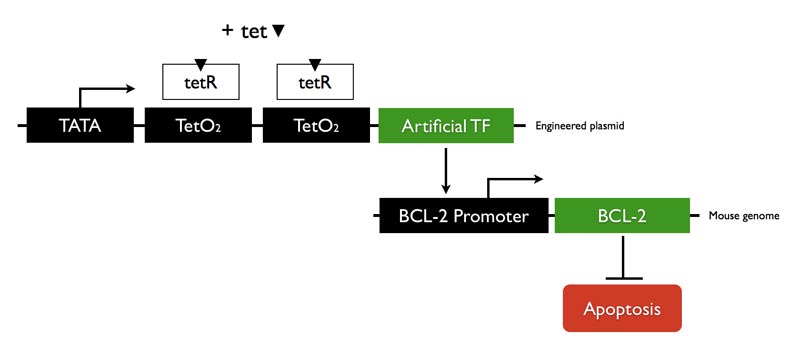
Using a zinc-finger Artificial Transcription Factor (ATF) backbone, we designed an in vivo gene regulation system of Bcl-2. This gene was chosen due to its transcription of an empirically important protein; it also offers a good system for conducting experiments in vivo through its involvement in cellular disease states, particularly in reference to cellular mitochondria. We prepared a highly developed ATF that is programmed to bind the promoter region of Bcl-2 and is subject to control by tetracycline.
Experiment
We tested our construct for function in mammalian cells. The experiments were conducted by introducing our ATF into mouse fibroblast cells, adding tetracycline in certain conditions, and then measuring protein levels made by the cells. Our project design predicts that cells that have the ATF and receive a dose of tetracycline should have increased Bcl-2 gene expression. We would then be able to detect this up-regulated gene activity by measuring the subsequent increase production of Bcl-2 protein by western blot.