Lethbridge/Summary
From 2007.igem.org
Lethbridge iGEM Project Overview
The goal of our project is to create a ‘bio-prospector;’ a microorganism capable of moving up a concentration gradient for a particular small molecule (theophylline), and activating either a metabolic or signaling pathway. We hope that once this system is developed, it will be easy to modify it to be specific to many different small molecules. To achieve this, we will use a riboswitch ([http://pubs.acs.org/cgi-bin/abstract.cgi/jacsat/2007/129/i21/abs/ja0692480.html Topp and Gallivan, 2007]) to control the levels of CheZ, a protein involved in motility.
E. coli can travel in their environment by sensing chemical signals. Normally E. coli sense these chemicals using their five transmembrane receptor proteins. These proteins interact with the flagellar motor via a well characterized set of cytosolic proteins (CheA, B, R, W, Y, and Z). In the presence of an appropriate ligand, CheA is autophosphorylated. This phosphate group is transferred to CheY, which then interacts with the motor complex. When this occurs, the motor rotates clockwise, and the E. coli cells tumble. CheZ dephosphorylates CheY, causing the motor to rotate counterclockwise, and the cells to swim straight (a 'run'). In CheZ knockout strains, the E. coli cells are in a constant tumbling state. Introducing CheZ into these mutant strains, and controlling its expression would allow us to control the movement of the organisms.
Our project will involve attaching a riboswitch in front of CheZ, that in the ‘off’ state will repress translation of CheZ, while in the ‘on’ state will allow its translation. This control would be happening at the mRNA level. A riboswitch is an RNA construct that resides in the 5' UTR of the gene it regulates. It contains an aptamer that will assume different conformations in the presence or absence of a particular ligand. One of these conformations will allow ribosome binding – and hence protein synthesis – while the other will inhibit ribosome binding, usually through physical interaction with the ribosome binding site. In our system, when the ligand (theophylline) is present, the ribosome can bind, and translation occurs. After translation of CheZ occurs, CheY can be dephosphorylated, and the E. coli cells will swim normally.
As ligand concentration increases, the incidence of ligand-riboswitch binding and resultant translational activation of CheZ will increase. This will cause the cell to move a higher percentage of the time resulting in a net movement up the concentration gradient.
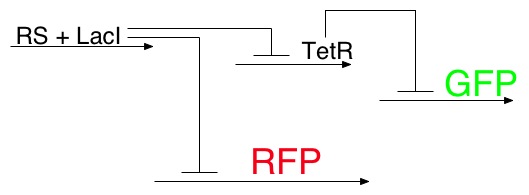
To determine the concentration of the ligand, we use a color output similar to many other projects, with one twist. The concentration of the lead signal, LacI, will also be controlled by a riboswitch. Thus the more ligand present, the higher the levels of LacI, which in turn will determine which phosphorescent protein(s) will be produced.
The overall approach will be two-fold. The first aspect will be to start combining the components required to synthesize our system (in parallel) to ensure that the components interact properly and as expected. Secondly, simulations will be done to determine what alterations (such as degration flags or activation sites), if any, are required to make get the best results from the system.
The nature of both the riboswitch and the iGEM biobrick method fit hand in hand. Thus our long term goal is to expand the number of riboswitches and to combine them with existing bricks to create novel systems.
Chemotaxis System
Reporter System
RS=Riboswitch
Main Page | Project Overview | Timeline | Lab Notebook | Team Members