Virginia/Projects/1
From 2007.igem.org
| HOME | PROJECT INTRO | APPROACH | PROCEDURES | RESULTS | [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2007&group=Virginia BIOBRICKS] | [http://openwetware.org/wiki/IGEM:VGEM/2007/Notebook eNOTEBOOK] | [http://www.seas.virginia.edu/VGEM/ WEBSITE] |
Harvesting Biomass and Light to Power Butanol Biosynthesis
At the 2007 iGEM Jamboree, we will present our synthetic biology approach to the design and implementation of a novel metabolic pathway in E. coli that utilizes cellulose and sunlight as sole energy sources for the biosynthesis of butanol, an alternative liquid transportation fuel. Click [http://www.openwetware.org/wiki/IGEM:VGEM/2007/Projects here] to find our more about our other projects that will not be presented this year.
Approach
We have designed a metabolic system capable of cellulose degradation and light metabolism in order to power the biosynthesis of butanol fuel. This hybrid molecular engine is built from standard biological parts and may be used in future designs in order to drive forward cellular chemistry.
The coming years are going to require us to revamp our notions of a fuel economy. Our team hopes to show how synthetic biology can aid in tackling real-world problems not only to aid in the development of new fuel technologies, but also to help support the synthetic biology community by showing it's utility.
Our Approach
Our goal is to isolate the pathway of butanol production existing in various organisms and engineering the metabolic pathway of E.Coli to produce butanol. Butanol limits bacterial growth by degrades cellular membranes, so the first step is to convey butanol tolerance to E.Coli. This will be accomplished via the use of tolerance genes from other bacterial species.
Next, we will transform the cells with the necessary enzymes for butanol production. These are explained in detail below. By growing the cells in anaerobic conditions and analyzing their product, we hope to tweak the pathway to produce maximum amounts of butanol.
One approach to this is to vary the energy sources the bacteria can use. By inserting genes coding for cellulase, we hope to give our cells the ability to use cellulose as an energy source. Agricultural waste would then become the feed for our strains. Additionally, the use of proteorhodopsin to supplement ATP production is planned. Proteorhodopsin allow the cells to harness light energy independent of oxygen in the environment and drive cellular metabolism.
Our final goal is to design a system that will allow E.Coli to be tolerant to butanol, produce butanol, and do so by exploiting various energy sources to increase efficiency and large-scale feasibility.
Biobrick and pathway design
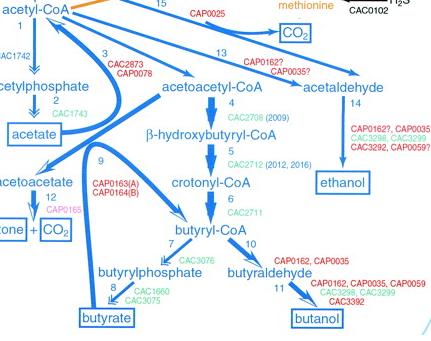
Image source: http://jb.asm.org/cgi/content/full/183/16/4823/F5
This is the pathway we are incorporating into E.Coli.
Clostridium acetbutylicum has been known to produce butanol anaerobically in nature. For our project we will be utilizing two butanol sythesis pathways available to us from its genome. In addition to this, we plan to provide a carbon source for butanol biosythesis by importing a cellulase gene from Saccaruphagus degradans, and we plan to facilitate the proton gradient required for ATP sythesis through the use of a proteorhodopsin system.
The diagram below depicts the complete system that we plan to create. The added pathways are: 3, 4, 5, 6, 9, 10, 11. Excluded from this image is our plans for the cellulose digestion pathway, which essentially feed into the production of pyruvate.
The specific proteins we need the cells to produce for this pathway are:
- Specific cellulase
- butanol tolerance genes
- thiolase
- beta-hydroxybutyryl-CoA dehydrogenase
- crotonase
- butyryl coa dehydrogenase
- AAD/AAD2
- alcohol dehydrogenase
- AOTC/AOTD
We have designed the following biobricks:
Need this info.
Send comments to George McArthur