ETHZ/Engineering
From 2007.igem.org

.:: Introduction ::.
In order to understand if it is possible to create the learning system that we wanted, we had to run some initial simulations, to see if we could reach the desirable steady states. After creating a basic framework on which to work on, we refined the parameters by searching the available literatures. In the next, we are presented the coupled differential equations that model our system, their parameters and the values that we picked, the results of our simulations, and lastly, we provide our references.
.:: System Model ::.
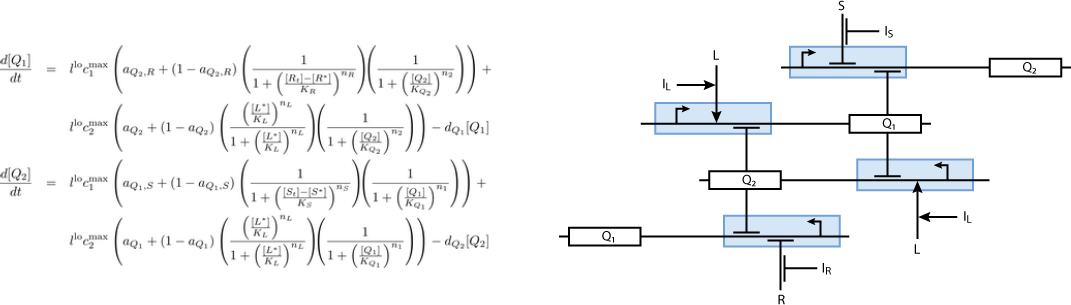
Based on [1], we modeled the biological system with differential equations. According to what presented in the Biology Perspective, our system is composed of three subsystems:
- A subsystem of constitutively produced proteins,
- The learning part, and
- The reporting subsystem.
The subsystem with the constitutively produced proteins serves as a regulatory system, and can be modeled with three decoupled partial differential equations:
The second subsystem is the main part of the biological model. This subsystem stores the information concerned the learned chemical, and drives the production of the appropriate reporter, during the recognition phase. It is actually a toggle switch, that reached a steady state depending on the chemical that it is exposed to:
.:: Simulations ::.
.:: References ::.
[1] This book
[2] A synthetic time-delay circuit in mammalian cells and mice (http://www.pnas.org/cgi/content/abstract/104/8/2643)
[3] Detailed map of a cis-regulatory input function (http://www.pnas.org/cgi/content/full/100/13/7702?ck=nck)
[4] Parameter Estimation for two synthetic gene networks (http://ieeexplore.ieee.org/iel5/9711/30654/01416417.pdf)
[5] Supplementary on-line information for "A Synthetic gene-metabolic oscillator" (no link)
[6] Genetic network driven control of PHBV copolymer composition (http://doi:10.1016/j.jbiotec.2005.08.030)