Edinburgh/DivisionPopper/Status
From 2007.igem.org
MENU : Introduction | Background | Applications | Design&Implementation | Modelling | Wet Lab | Synthetic Biology Approach | Conclusions
Contents |
Plan to model the system:
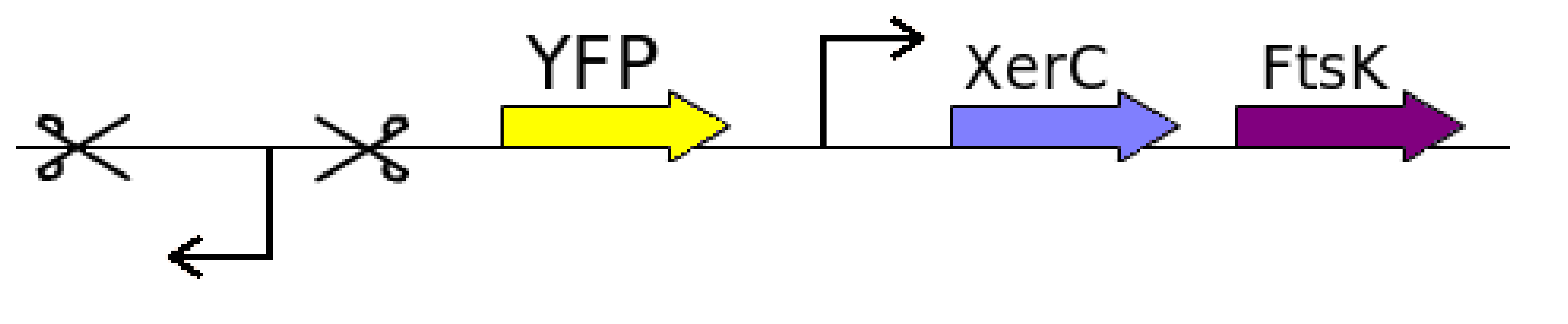
It is always good to move stepwise instead of jumping into the final idea. To show that the diff sites actually flips every cell division, we started with the proof of concept. The proof of concept involves constructing a construct with a reverse Lac promoter in between diff sites followed by YFP gene next to diff-rev Lac P-diff. Ftsk and Xer were planned to be inserted as it was stated in a paper that extra production of Xer and Fts apart from normal cell production will get better results. The following the proof of concept design:
Requirements:
1. A low copy number plasmid. Ideally only one plasmid should be present. 2. To check the cell changing to yellow colour after flip occurs by means of a fluorescent microscopy, we had to largely delay the doubling time of the cell. 3. The fluorescent protein should be degraded completely before the cell division occurs.
Precautions taken to satisfy the requirements:
1. We initially searched for minichromosomes as we found that the oriC was included in the plasmid as a factor to make the plasmid replicate like a chromosome. But the literature search led us to the finding that every cell has 15 copies. So we searched for miniF plasmid with very low copy number. We found () to have a copy number of 1-2. We got the plasmid from Dr.Masters, University of Edinburgh and used it for building the construct. 2. To delay the doubling time we decided to use minimal medium which would delay the division to more than an hour Medium: 3. In order to account for the fluorescent proteins rapid degradation, a rapid degradation tag (AANDENYALVA) was tagged to the fluorescent protein gene by including the tag in the primer sequence so that the amplified PCR product will have the gene with the rapid degradation tag. This tag with the LVA increases the degradation rate to the maximum ( -0.018 1/min) which actually brings down the half life of the protein to approximately 40 min.( Thanks for the paper Dr.Drew Endy)
Construct building:
All the necessary parts were taken from the registry. The oligonucleotides for reverse and forward dif-sites (difF and difR) were amplified, combined and biobricked.
Primers for a reverse lac promoter (PlacRev) were ordered and the sequence amplified and biobricked.
This reverse lac promoter was combined with the dif forward sequence to give the biobrick difF-PlacRev.
Yellow fluorescent protein (YFP) was revived from the Registry and combined with the reverse dif-site to yield the biobrick difR-YFP, where YFP is the reporter gene.
A control sequence that consists of the lac promoter and YFP was made for imaging purposes.
A miniF plasmid was transformed and plated. Primers for FtsK and XerC were ordered and said sequences amplified, digested and biobricked, but only FtsK yielded a final biobrick; cells overexpressing XerC refused to grow.
General Procedure involved:
The general procedures involved in the process of biobricking are as follows:
- Design forward and reverse primers for the sequence of choice
- PCR amplify the sequence with said primers
- Double restricion digest the PCR amplified sequence and a vector (Edinbrick1) to create dissimilar sticky ends
- Purify the reactions
- Mix cut vector and insert with DNA ligase overnight
- Transform the biobrick into a chemocompetent E. coli cell line
- Plate high and low concentrations of transformed cells onto selective plates and leave overnight
- Replate colonies and inoculate broths overnight with antibiotics that select for the vector
- MiniPrep broths that correspond to successful replated colonies
- Do analytical restriction digest and/or PCR reactions and run samples on agarose gel to check the insert size
- Sequence the produced construct.
- MaxiPrep correct biobricks.
It goes without saying that trial-and-error preceeded optimal PCR conditions for each designed primer. We have also employed high-resolution agarose gels when required.
Three attempts to combine difF-PlacRev and difR-YFP have failed to date. We have employed a selection of techniques including gel band excision and purification of both vector and insert and differential restriction enzyme and ligation times. Lab work continues.
Up next is fluorescence microscopy of the control and final construct.
We're eagerly anticipating the arrival of the more advanced construct (the actual division PoPper) via GeneArt, but their email correspodence is not promising. The word is that our orders are heavily Delayed. The clock is ticking...
Biobricks
For the construction of experiments and the Division PoPper several Biobricks are needed. Some of them have been taken from the Registry of Standard Parts and some have been constructed from scratch.
A total of thirteen biobricks were designed, details of which you'll find on the [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2007&group=Edinburgh Registry team page].
Biobricks
For the construction of experiments and the Division PoPper several Biobricks are needed. Some of them have been taken from the Registry of Standard Parts and some have been constructed from scratch.
A total of thirteen biobricks were designed, details of which you'll find on the [http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2007&group=Edinburgh Registry team page].