Cambridge/Signalling project
From 2007.igem.org
One part of our project involved implementing a new signalling mechanism for communication between distantly-related bacteria (in particular the Gram-negative E. coli and the Gram-positive B. subtilis. The natural system on which we based our attempt was the Agr peptide quorum-sensor from S. aureus. This page describes the natural system, the steps taken to convert it into a BioBrick-compatible form and results of experiments on the constructs produced.
Contents |
Introduction
Presently, the world of synthetic biology is critically short of robust inter-cellular signalling systems needed to engineer scalable applications (since a single cell can only ever accomplish a limited part of an overall function). The main system in current use is the lux system from the Gram-negative Vibrio fischeri (sender BioBrick F1610, receiver BioBricks F2620, F2621, F2622). This system is based on a homoserine lactone (HSL) compound - although other HSL-based systems exist, there are problems with crosstalk between them.
In contrast, in the Gram-positive world there are many oligopeptide-based signalling systems (usually used for quorum sensing in bacterial populations) that have been well studied and shown to have low crosstalk levels. In this subproject we aimed to convert one of these, the agr system from S. aureus, into a usable signalling device as a paradigm for many more communication channels in future.
Natural system
Background
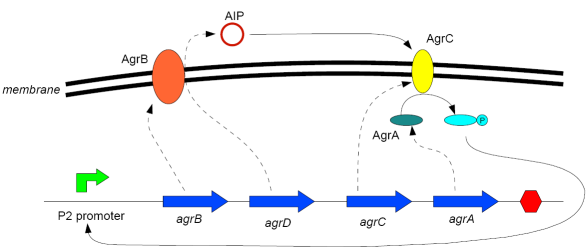
The Accessory Gene Regulator (agr) locus is responsible for quorum sensing and control of virulence in Staphylococcus aureus. It is involved with both the secretion and the detection of a short polypeptide, modified post-transcription to produce a thiolactone ring; this molecule has been termed AIP (AutoInducing Peptide). AIP serves a quorum-sensing role for S. aureus, in that when the population density is sufficient (as measured by AIP concentration) the bacteria activate virulence genes and begin to cause adverse effects to their host (Novick et al, 1993; Ji et al, 1995).
Genetics
The agr locus contains two divergent transcription units, denoted 2 (or II) and 3 (or III), under the control of promoters P2 and P3 (Peng et al, 1988). RNAII, under the control of the promoter P2, contains four open reading frames - in order of transcription:
- agrB: A membrane protein that acts as a synthetase for AIP
- agrD: A relatively short polypeptide that is the precursor of AIP (cleaved and modified by AgrB)
- agrC: A membrane protein that acts as a sensor for AIP
- agrA: A response regulator that is phosphorylated by AgrC when AIP is bound to it, and then in turn binds to the promoters P2 and P3 to enhance their transcription.
Diagram:
(after Waters and Bassler, 2005)
The RNAII transcript is thus "doubly autocatalytic" (Ji et al, 1995) - the presence of AIP both increases the rate at which AIP is made and also makes the cell more sensitive to the oligopeptide.
RNAIII, transcribed from unit 3 under the control of P3, activates virulence genes elsewhere in the cell once the AIP concentration reaches a certain level and is not of interest for this project.
AIP variants
To date 4 different varieties of AIP have been identified; these involve differences both at the gene loci and at the level of the signalling molecule. The different types are not only incompatible but, for the main part, cross-inhibitory (Ji et al, 1997; McDowell et al, 2001).
We selected Agr group I, originally from S. aureus strain RN6390B, as the sequence for this was readily available (see below).
DNA sequence data
Strain RN6390B is a derivative of strain NCTC8325 (Peng et al, 1988; NARSA A), which has been sequenced by (Roe et al A). An annotated version was downloaded from (NCBI Entrez Genome A); the agr coding regions are the ORFs labelled SAOUHSC_0226{1,2,4,5}, running from 2,093,874bp to 2,096,632bp. The non-coding P2 promoter region of the sequence is less well defined. The overall extent of the promoter region is indeicated in (Morfeldt et al, 1996), (Rao et al, 1995) shows potential the transcription start sites and (Koenig et al, 2004) indicates the areas of the sequence to which phosphorylated AgrA binds. Shine-Dalgarno ribosome binding sequences were manually identified.
Based on the above, [http://www.ccbi.cam.ac.uk/iGEM2007/images/9/9e/Agr_operon_annotated.gb an annotated section of the (Roe et al A) genome sequence containing the agr operon] was produced in GenBank format, with labelled promoter regions, putative RBSes and coding regions.
Synthetic sender and receiver constructs
The sequences for agr{A, B, C, D} and the P2 promoter, described above, were used to design three BioBricks (MIT Registry of Standard Biological Parts A) corresponding to the three sections of the sensing system. This involved codon optimisation (using the GeneDesigner software package (Villalobos et al, 2006; DNA2.0 A)) and the addition of the BioBrick prefix and suffix sequences to allow standard assembly. Additional restriction enzyme sites were also inserted between coding regions to allow future disassembly and reconfiguration of the devices.
In principle, due to the similarity between S. aureus and other Gram-positive bacteria such as B. subtilis, the components of the system will function correctly when transferred to such a chassis. In order to use the system in Gram-negative bacteria such as E. coli, additional measures were necessary due to the presence of the outer membrane; see below for details.
Simple AIP sender device
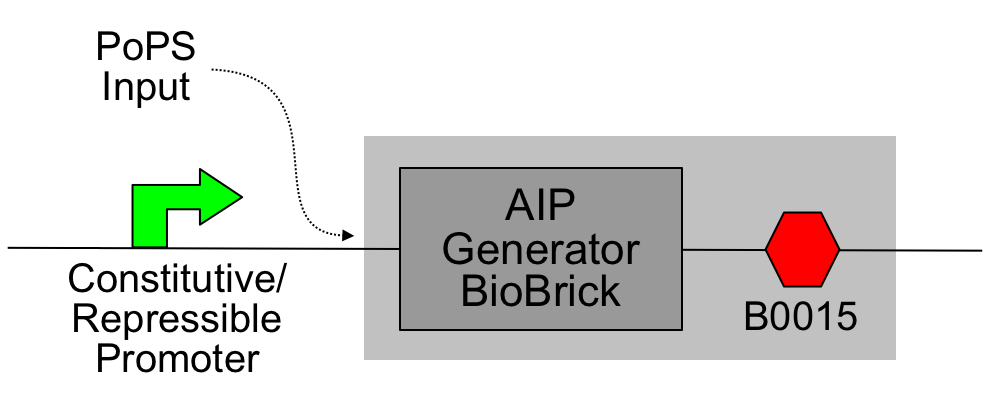
The core of this device is the "AIP Generator BioBrick" ([http://partsregistry.org/Part:BBa_I746000 I746000]):
(download the sequence in [http://www.ccbi.cam.ac.uk/iGEM2007/images/a/a4/AIP_generator_biobrick.fa Fasta format] or [http://www.ccbi.cam.ac.uk/iGEM2007/images/9/9e/AIP_generator_biobrick.gd GeneDesigner format])
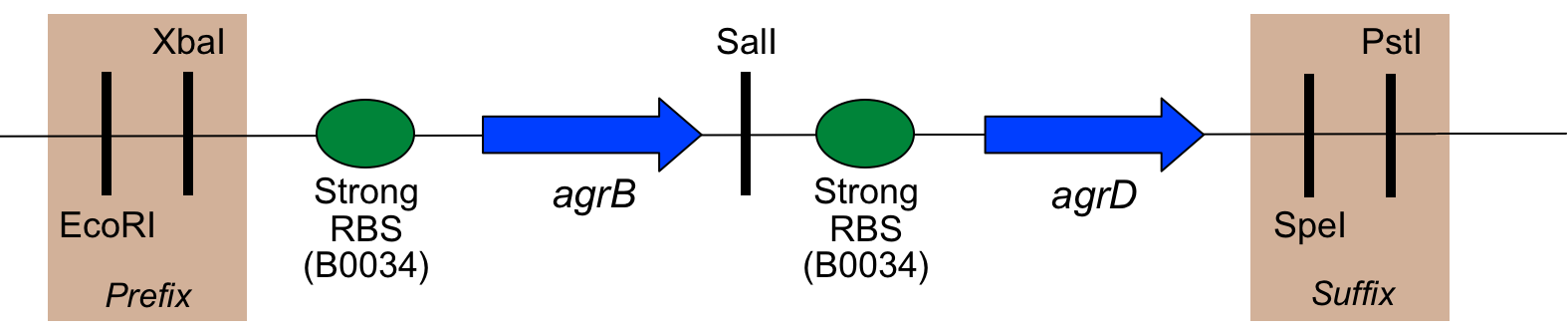
As mentioned above, several changes were made to the natural sequence of the agr coding regions to produce this part:
- Codon usage has been optimised for E. coli (which also seems to give good results for B. subtilis
- The start codon of agrB has been changed from TTG to ATG
- A SalI restriction site has been added between agrB and the RBS before agrD
- Biobrick "scars" between components, as well as a prefix and suffix, have been added
This was then assembled with the [http://partsregistry.org/Part:BBa_B0015 B0015 terminator] to produce a PoPS->AIP device ([http://partsregistry.org/Part:BBa_I746001 I746001]); constitutive promoters from the [http://partsregistry.org/Part:BBa_J23100 J231XX series] were added to produce constitutive sender cells.
Schematic:
The light blue shading indicates the device; the constitutive promoter may be omitted and the PoPS input derived from an earlier stage of logic.
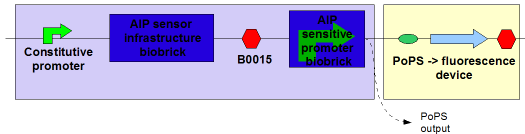
Simple AIP receiver device
This device contains two new BioBricks, the "AIP Sensor Infrastructure BioBrick" ([http://partsregistry.org/Part:BBa_I746100 I746100]):
(download the sequence in [http://www.ccbi.cam.ac.uk/iGEM2007/images/7/74/AIP_sensor_infrastructure_biobrick.fa Fasta format] or [http://www.ccbi.cam.ac.uk/iGEM2007/images/7/75/AIP_sensor_infrastructure_biobrick.gd GeneDesigner format])
And the P2 promoter, sensitive to phosphorylated AgrA ([http://partsregistry.org/Part:BBa_I746104 I746104]):
(download the sequence in [http://www.ccbi.cam.ac.uk/iGEM2007/images/d/dd/AIP_sensitive_promoter_biobrick.fa Fasta format] or [http://www.ccbi.cam.ac.uk/iGEM2007/images/4/4f/AIP_sensitive_promoter_biobrick.gd GeneDesigner format])
Similarly to the AIP generator device, some changes have been made to the natural sequence:
- Codon usage has been optimised for E. coli (which also seems to give good results for B. subtilis)
- A BamHI restriction site has been added between agrC and the RBS before agrA in the AIP Sensor Infrastructure BioBrick
- Biobrick "scars" between components, prefixes and suffixes have been added
Together these were assembled (via a number of intermediates, using the [http://partsregistry.org/Part:BBa_B0015 B0015 terminator] and constitutive promoters from the [http://partsregistry.org/Part:BBa_J23100 J231XX series]) into an AIP->PoPS transducer:
The light blue shading indicates the device. The constitutive promoter causes the production of the components of the signal transduction pathway (AgrC and AgrA); when phosphorylated AgrA is present, the AIP-sensitive promoter is activated and PoPS is output. In this setup a fluorescent protein is produced (GFP from [http://partsregistry.org/Part:BBa_E0840 E0840]), but in an application this would feed into a downstream device.
Importing the system into E. coli
It was hoped that the Bacillus subtilis chassis would be ready in time for testing of the AIP system. However it was also thought prudent to investigate a means for using the system in E. coli in case circumstances prevented such a test.
The problem with importing the system into E. coli is the presence of the outer membrane in Gram-negative bacteria. However, it was believed that this would be solved by introducing a protein pore into the outer membrane to allow AIP to diffuse out of periplasm of sender cells and into the periplasm of receiver cells. (This is all the extra machinery that is necessary, since the accepted model suggests that AgrB exports AIP through the inner membrane and AgrC is present in the outer membrane to detect AIP in the periplasm.)
Such a protein pore was found in the form of a mutant of the FepA siderophore transport protein, produced during the research described in (Newton et al, 1999). The delL8 and delL8T mutants in particular seem (from the conclusions reached in the paper) to be ungated open channels, with the demonstrated ability to admit large antibiotics such as [http://en.wikipedia.org/wiki/Bacitracin bacitracin]; since this is a cyclised oligopeptide with a molecular mass of 1400 Da (AIP is around 900Da), it is a positive indication that FepA may be permeable to AIP.
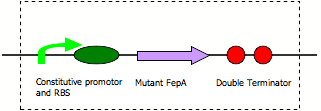
FepA permeability device
Diagram:
BioBricking of FepA cDNA
We were initially supplied with the delL8 and delL8T mutants of FepA described in (Newton et al, 1999). The coding sequences were on the pUC19-derived plasmid pITS449, under the control of their natural fur-regulated promoter, and hosted in E. coli strain KDF541.
The first task was to PCR out the cDNA for the mutant proteins alone, with the BioBrick ends attached. An added difficulty was the presence of an EcoRI site within the coding region, which had to be removed to allow compatibility with the BioBrick standard. This was achieved by site-directed mutagenesis with a "megaprimer"-based method, using two PCR stages for each mutant. The first stage used a forward primer (with BioBrick prefix) and an internal mutagenic primer designed to remove the EcoR1 site via a silent mutation. The product of this PCR reaction, the so-called "megaprimer", was then used in combination with a reverse primer and the original template DNA in a second PCR reaction to generate the complete mutated sequence.
Standard Construction of Permeability Device
The first standard construction was with the usual Elowitz Ribosome Binding Site ([http://partsregistry.org/Part:BBa_B0034 B0034]). The intermediates generated were then assembled as inserts, in front of a terminator ([http://partsregistry.org/Part:BBa_B0015 B0015]). After this, promoters were added. Since it was not known what level of FepA expression was required to confer sufficient permeability and whether too much expression would cause a significant growth defect, a range of constitutive promoters with different strengths from the [http://partsregistry.org/Part:BBa_J23100 J231XX series] was used. The promoters used were: [http://partsregistry.org/Part:BBa_J23100 J23100], [http://partsregistry.org/Part:BBa_J23111 J23111], [http://partsregistry.org/Part:BBa_J23107 J23107] and [http://partsregistry.org/Part:BBa_J23115 J23115]. These constructions were not wholly successful first time. All of the constructs involving the delL8 mutant worked, but only the J23115 delL8T ligation gave transformants.
Testing and Troubleshooting the Permeability Device
Since most of the constructs were complete, it was decided to press ahead with testing despite the failures. Having, no source of AIP and no way to assay for it, the best indication of permeability available to us was to repeat the bacitracin assay described in (Newton et al, 1999). 1.5ml of overnight cell culture was mixed with 1.5ml of 1.4% LA and poured onto a plate to give a lawn of bacteria. Sterile filter paper discs were placed onto the agar after drying and then spotted with 5ul of 2U/ul bacitracin. Inhibition of growth around the disc shows bacitracin permeability, since E. coli are not normally sensitive to this antibiotic. Unfortunately, on the first round of testing, none of the constructs seemed to confer permeability.
At this point two of the constructs were sent for sequencing, one representing each mutant. It was found that the delL8 mutant had a significant deletion in an unexpected region of the protein whereas the delL8T mutant sequence was correct. The failed constructions were therefore redone, and cells were retransformed with DNA from the one correct construct. The assay was also altered, this time using only 100ul of cell culture with 3ml of 0.7% LA. With this altered assay, the already completed construct (now called [http://partsregistry.org/Part:BBa_S03797 S03797]) did give a positive result. After further construction attempts two more constructs were also completed, [http://partsregistry.org/Part:BBa_I746201 I746201] and [http://partsregistry.org/Part:BBa_I746202 I746202], and these too showed bacitracin sensitivity.
This image shows the zone of clearing around a bacitracin-soaked disc. The part being expressed here is I746202, but other parts showed similar diameters of clearing. This was seemingly unrelated to promoter strength.
AIP sender device with permeability
For these constructs, the simple AIP sender device described above ([http://partsregistry.org/Part:BBa_I746001 I746001]) was inserted upstream of two different FepA permeability devices (I746201 and S03797), producing [http://partsregistry.org/Part:BBa_I746009 I746009] and [http://partsregistry.org/Part:BBa_I746010 I746010] respectively. These constructs were then ligated in front of various constitutive upstream promoters. Only constructs with the AIP controlled by the weakest promoter used (J23115) seemed to grow ([http://partsregistry.org/Part:BBa_I746210 I746210] and [http://partsregistry.org/Part:BBa_I746211 I746211] respectively). Even then, the construct with the weaker permeability device (I746211) grew very strangely in liquid culture, forming large clumps instead of an even cell suspension.
AIP receiver device with permeability
For these, the partial AIP receiver device with a PoPS->GFP transducer was assembled before the permeability devices (I746201 and S03797). Problems with assembly meant that only the former construction was taken forward to be assembled with J23107, providing a constitutive promoter to complete the receiver device ([http://partsregistry.org/Part:BBa_I746220 I746220]).
Testing of E. coli senders and receivers
No suitable assay could be found for the detection of AIP by non-biological means and it could not be sourced in pure form, so complete system tests were carried out by combining the senders (I746210) and receivers (I746220) and attempting to detect increased production of GFP.
This was done in several ways:
- Firstly, overnight cultures of receivers were diluted 1:100, grown for 2 hours and then induced with varying amounts of supernatant from overnight sender cultures. They were viewed after two hours on a fluorescence microscope.
- A variation on this was to induce the receivers with sender supernatant and then use a plate reader to measure fluorescence over time.
- Finally, a lawn of receivers was prepared by diluting 100ul of culture into 3ml of molten top agar, and induced by spotting supernatant or raw sender cell culture onto a filter paper disc in the centre of the lawn.
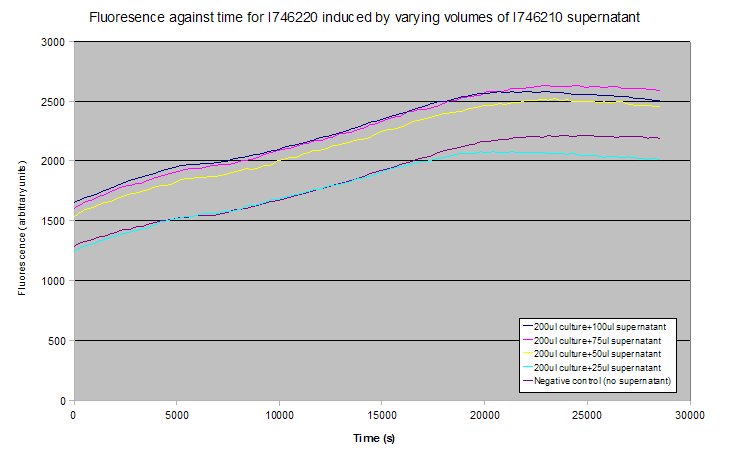
None of these tests showed induction - results for the plate reader experiment are shown below (averages of 3 experiments). Notice that the gradients of the fluorescence-time curves relate to the rate of transcription of GFP, and these are approximately constant between all the curves despite their relative vertical displacement.
The lack of induction could have been due to a missing, uncharacterised repressive element that is in S. aureus but not present in E. coli, as uninduced controls still showed a significant level of fluorescence. Alternatively, AIP or one of the Agr system transcripts might not have been folding properly, might have been aggregating, or might not pass through the FepA pore.
Conclusions
Achievements
This subproject attempted to implement a peptide signalling system in a Synthetic Biology framework, as a paradigm for multi-channel parallel interference-free intercellular communication. Unfortunately we were unsuccessful in this aim - the system was constructed but initial tests indicate a failure within it and time has not allowed us to identify and correct the fault.
However, a side aim of the project - that of introducing porosity for large molecules into the outer membrane of E. coli - has succeeded, with the contribution of a number of novel BioBricks to the Registry.
Above all, the subproject has demonstrated the context-dependence of the functioning of cellular mechanisms and the difficulties with fault-finding in biological systems.
Potential for further work
- Debugging the Agr signalling system: A number of steps could be undertaken to identify the fault currently preventing the system from working:
- Using HPLC and mass spectrometry, or a biological assay with S. aureus, to test for the production of AIP by the sender cells.
- Testing for the presence of the Agr{A, B, C, D} polypeptides.
- Import into eukaryotic cells: In principle, peptide signalling systems may also be used for communication between eukaryotic cells. A number of checks must be made before we can be confident these biobricks will work in either plant or mammalian systems.
- Checking for donor sites, acceptor sites or branch points for intron splicing. This may be done using [http://www.cbs.dtu.dk/services/NetPGene/ NetPlantGene].
- Checking for sites signalling downstream polyadenylation (the sequence AATAAA). None of the novel BioBricks described above contain this sequence (at all, let alone in a coding region).
It is hoped that by submitting the relevant constructs to the Registry other researchers will be able to take up this problem in future.
Acknowledgements
We are extremely grateful to DNA 2.0 Inc. (http://www.dna20.com) for sponsoring us by synthesising the AIP Generator BioBrick and AIP Sensor Infrastructure BioBrick described above.
References
- Novick, Ross, Projan, Kornblum, Kreiswirth and Moghazeh (1993) - Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. The EMBO Journal Vol. 12 no.10 pp.3967-3975
- Ji, Beavis and Novick (1995) - Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA Vol. 92, pp. 12055-12059, December 1995, http://www.pnas.org/cgi/reprint/92/26/12055.pdf?ck=nck
- Peng, Novick, Kreiswirth, Kornblum and Schlievert (1988) - Cloning, Characterization, and Sequencing of an Accessory Gene Regulator (agr) in Staphylococcus aureus. J. Bacteriol., 179, 4365-4372
- Waters and Bassler (2005) - Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005 21:319–46, http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.cellbio.21.012704.131001
- Ji, Beavis and Novick (1997) - Bacterial Interference Caused by Autoinducing Peptide Variants. Science 276 (5321), 2027, 27 June 1997
- Philip MDowell, Zina Affas, Caroline Reynolds, Matthew T. G. Holden, Stewart J. Wood, Sandra Saint, Alan Cockayne, Philip J. Hill, Christine E. R. Dodd, Barrie W. Bycroft, Weng C. Chan, Paul Williams (2001) - Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Molecular Microbiology (2001) 41(2), 503–512, http://www.blackwell-synergy.com/doi/abs/10.1046/j.1365-2958.2001.02539.x
- Lyona and Novick - Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria
- NARSA (A) - http://ww.narsa.net/control/member/search?repositoryId=105
- B.A. Roe, Y. R. Tian, H. Jia, S. Li, S. Lin, S. Kenton, H. Lai, JD White, A. Dorman, F. Z. Najar, S. Clifton, V. Worrell and J. Iandolo (A) - Staphylococcus aureus Genome Sequencing Project, http://www.genome.ou.edu/staph.html (direct link to sequence data: ftp://ftp.genome.ou.edu/pub/staph/staph-2k.fa)
- NCBI Entrez Genome (A) - accession number CP000253, http://www.ncbi.nlm.nih.gov/sites/entrez?db=genome&cmd=Retrieve&dopt=Overview&list_uids=19213 (direct link to sequence data: http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=CP000253 (untick the two "hide" checkboxes))
- Koenig, Robbin L., Ray, Jessica L., Maleki, Soheila J., Smeltzer, Mark S., Hurlburt, Barry K. (2004) - Staphylococcus aureus AgrA Binding to the RNAIII-agr Regulatory Region. J. Bacteriol. 2004 186: 7549-7555, http://jb.asm.org/cgi/content/full/186/22/7549
- L Rao, R K Karls, and M J Betley (1995) - In vitro transcription of pathogenesis-related genes by purified RNA polymerase from Staphylococcus aureus. Bacteriol. 1995 May; 177(10): 2609–2614, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=176928
- Morfeldt, E., K. Tegmark, and S. Arvidson (1996) - Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227–1237
- Salete M. C Newton, John D Igo, Daniel C Scott, Phillip E Klebba (1999): "Effect of loop deletions on the binding and transport of ferric enterobactin by FepA". Molecular Microbiology 32 (6), 1153–1165 - http://www.blackwell-synergy.com/doi/abs/10.1046/j.1365-2958.1999.01424.x
- MIT Registry of Standard Biological Parts (A) - http://partsregistry.org
- Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S (2006): "Gene Designer: a synthetic biology tool for constructing artificial DNA segments". BMC Bioinformatics. 2006 Jun 6;7:285.
- DNA2.0 (A) - http://www.dna20.com/index.php?pageID=78