McGill/Results
From 2007.igem.org
This year our new team has been developing the project established last year which you can see on the McGill 2006 wiki. We have made many new discoveries and have run into some problems hindering our progress along the way; nevertheless, we were able to make some new developments as outlined below:
Contents |
Reproducing Last Year's System
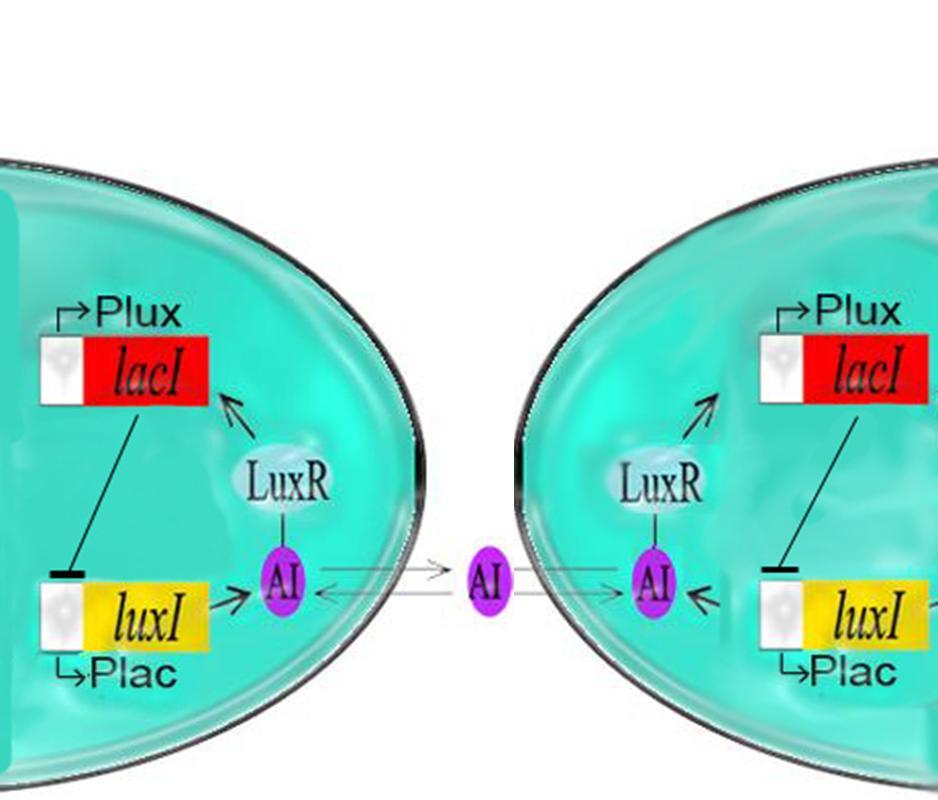
The primary oscillating system, the I15004 with the J40001 as outlined in the previous pages was produced last year and viewed using a microscope for several hours to analyze the oscillatory period and confirm the presence of these oscillations. Click here for methodsFrom these experiments, we were able to come up with some interesting results.
1. The more concentrated the cells, the worse the apparent oscillations.
2. The presence of Dox seemed to stabilize the oscillations.
3. The greater the amount of AHL, the worse the apparent oscillations, as predicted with the simulations.
4. Using an optical density assay, it was found that the cells were in fact dying (or clumping) thus lowering the optical density throughout the testing period.
5. Oscillations occured with the J40001 (pLux-LacI-ECFP) alone. The reason for this could be some interaction with the expression of endogenous Lac in the BL21 cell line we used.
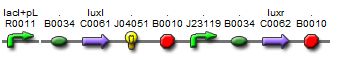
Production of a new I15004 biobrick
In light of the terrible bands following a restriction digest and the lack of oscillations in the MC cells, we constructed a new sequence of DNA containing the proper components of the I15004 needed for the oscillating network and added of few of our own.This sequence contains the following:
1. pLac, ribosome binding site, LuxI These three components are present from the I15004 and are required for the oscillating network.
2. RFP The RFP is our own addition. It allows us to track the expression of both the oscillating I15004 brick and the J40001 brick allowing us to evaluate oscillations in more than one way (note, a third method of reporting is included with the repressillator).
3. pConstitutive, ribosome binding site, LuxR Inculsion of this constitutive promoter was another one of our design ideas. When coupled with the repressilator, this eliminates one of the crossing links that would make the system more connected and create more variables. By eliminating the tetracycline promotor, our system is much cleaner.
As ideal and wonderful as this system sounds, we were unable to put it into action. We ordered the sequence from GenArt in early August and it has still not arrived in time for the Jamboree due to ligation problems at the company.
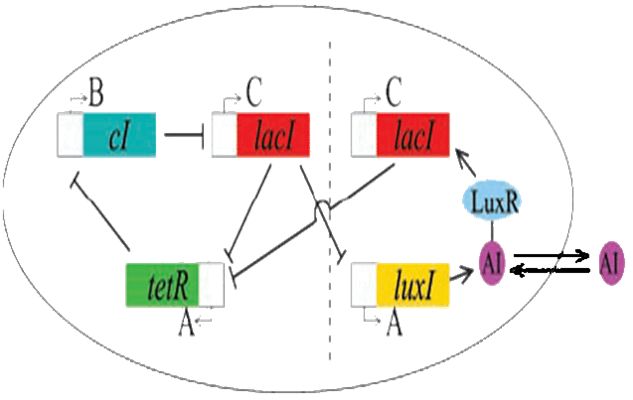
Coupling of the Oscillator with the Repressilator
As per the future prospects presented in 2006, one of the major goals of this year was to couple the oscillator with Elowitz's repressilator: a triple repressing system which will change the spiking oscillations of the dual oscillator to robust sinosodial curves.
Modelling using deterministic rate equations predicted beautiful curves that we have attempted to reproduce in the lab. Again, we ran into some problems. Repeated restriction digests produced anomalous gel results and when attempting to view the reporting gene in the repressilator, we were unable to see the fluorescence.
Our conclusion is that the I5610 is not what is appears to be. So, onto our next section : creating a new represillator. Thus, coupling was not yet possible in practice.