Princeton/overview
From 2007.igem.org
 Overview || People || Lab work || Experiments || Bioinformatics || Simulations
Overview || People || Lab work || Experiments || Bioinformatics || Simulations
Overview
2007: A Lentivirus-Delivered, RNAi-Enhanced Logic Circuit for Cancer-Specific Detection and Destruction
The Princeton University iGEM 2007 team, consisting of 10 undergraduate students, 2 high school students, and 8 instructors, envisions a paradigm shift in the way we envisage cancer cell targeting.
The majority of contemporary cancer treatments utilize rather unspecific approaches in targeting cancer cells. Such methods do not, and indeed cannot, specifically target a particular cancer cell type. As a result of their imprecision, these treatments tend to inflict almost as much damage upon healthy cells as cancerous ones. Such harmful side-effects often make cancer treatments such as chemotherapy or radiation therapy not only undesirable, but also less comprehensive, as their prolonged use is ultimately unsustainable. Our proposed, generalizable, system will resolve this unfortunate situation by enabling the detection and destruction of cancer cells in a tissue-specific manner.
Our system is engineered in such a fashion as to prevent any deleterious effects on non-cancerous cells or on cells that exhibit characteristics similar to cancer cells. To this end, RNA interference (RNAi) is employed to provide an additional level of regulation and prevent the apoptotic genes from being silenced by any epigenetic events. The added dimension of tissue-localization ensures that the system does not target any non-cancerous cell, thus, providing the most direct and effective cancer therapy.
We have selected breast cancer as our initial target. Breast cancer, according to the [http://www.who.int World Health Organization (WHO)] [http://www.who.int/mediacentre/factsheets/fs297/en/ (ref)], is the leading cause of cancer deaths in women worldwide. As a significant cause of cancer deaths, breast cancer serves as an appropriate cancer to target in a proof of concept implementation of our general design using the established MCF-7 cell line.
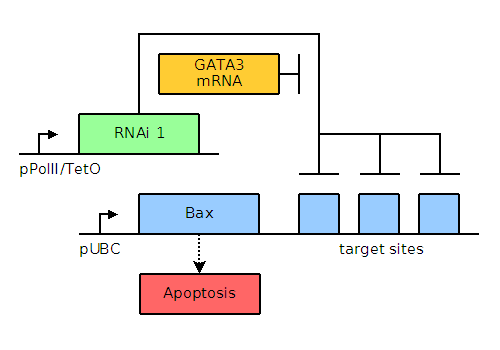
In MCF-7 breast cancer cells, GATA3, a non-neuronal ectoderm cell fate regulator and a transcription factor present in and important for the maintenance of breast tissue cells, is upregulated to levels approximately thirty-two times those found in healthy cells. When present in large quantities, as in the case of cancerous cells, GATA3 will titrate away the small interfering RNA (siRNA) and allow transcription and translation of the pro-apoptotic factors Bax or Bak. When present in small quantities, Bax and Bak are repressed by binding of the siRNA to the engineered stretch of sequences located either 5' or 3' to the Bax or Bak gene. A helper plasmid containing a mutated pol gene will enable the transient transfection of the entire system into the cancer cells, as opposed to the integration that would occur with the wild type pol gene. This transient effect will ensure that the apoptotic genes are not propagated by further cell division, thus preventing unintended proliferation of our construct.
Please see our lab work for current progress on our implementation.
Circuits
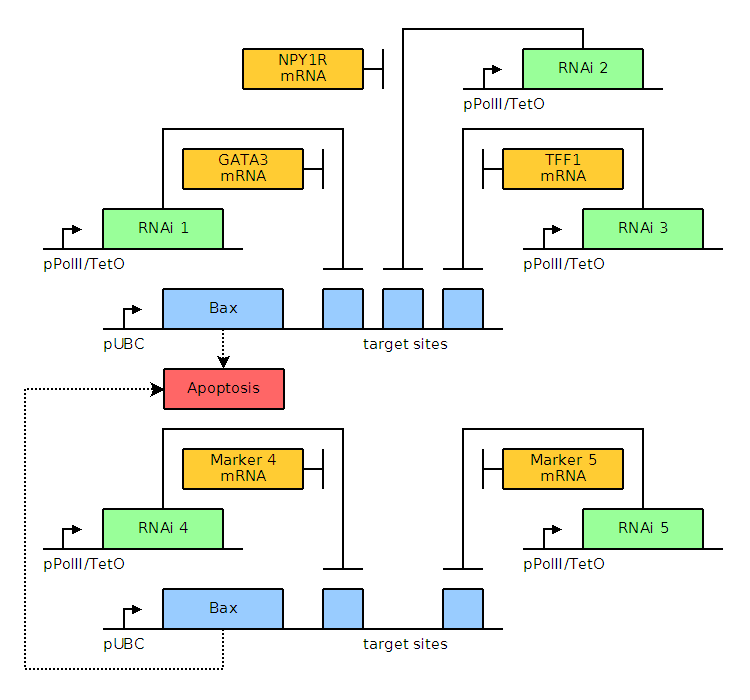
Through bioinformatics, it is noted that several factors, including GATA3, TFF1, and NPY1R, are overexpressed. The genetic circuits for systems of varying complexity are given below, including a basic system based on GATA3, an AND gate, and an AND OR gate. Blue and green elements are our constructs while orange elements are endogenous factors.