Duke/Projects/bp
From 2007.igem.org
Contents |
Bioplastics Synthesis
Background and Motivation
With rising concerns over environmental pollution and energy shortages, there has been a greater push towards alternative energy and environmentally friendly production of useful materials. Currently, the production of petrochemical plastics not only ejects hazardous gases into the atmosphere but also generates considerable amounts of waste. Along with a strong desire to become independent of oil-based products, these concerns have paved the way for research into highly biodegradable and biocompatible bioplastics, such as polyhydroxyalkanoates (PHAs), a particular polymer that is formed natively in several strains of bacteria. This project seeks to improve the production of a certain PHA, poly(3-hydroxybutyrate-co-4-hydroxybutyrate, through the maximization of the desirable 4-hydroxybutyrate monomer. This is because a high ratio of 3-hydroxybutyrate produces a very crystalline and brittle plastic while high 4-hydroxybutyrate produces a very desirable elastomers.
Parts and setup
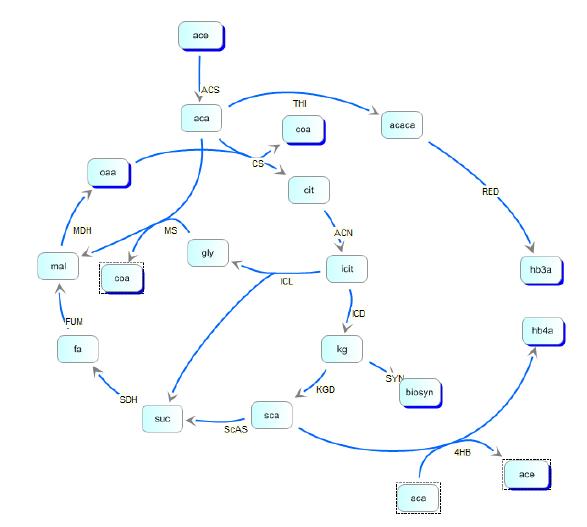
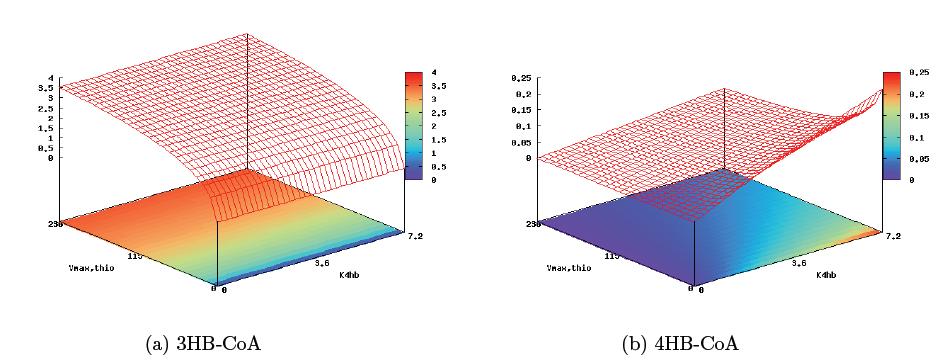
In order to optimize the experimental gene circuit and better control the ratio of 4-hydroxybutyrate (largely represented in the copolymer) to 3-hydroxybutyrate, we modeled the system in order to locate the limiting reaction steps in the metabolic pathway of E. coli and in the polymer producing extension of those pathways. With respect to the production of PHA, most of the modeling work have been black-box approaches focused on the optimization of PHAs in large industrial reactors. Another important aspect of PHA formation includes molecular weight distribution, which was treated in the work of Bradel and Mantzaris [21]. However, all these fractionated views of PHA synthesis failed to take into account the inherent metabolic coupling between PHA biosynthesis and other cellular processes. In the work of Iadevaia, et. al. the two were finally coupled in their treatment of a theoretical copolymer control of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) through the use of a genetic toggle switch. Kinetic pathways for 3HB and 4HB were developed and analyzed using integrated tools from the Systems Biology Workbench (SBW). Jdesigner2 was used to build the network and deterministic modeling and steady-state analysis were performed using Jarnac[24]. Reaction kinetics of the tricarboxyilic acid cycle were reconstructed from the work of Singh, et. al., while reaction rates and kinetics for 3HB kinetics were obtained from the work of Iadevaia, et. al. Various other miscellaneous enzyme rates were obtained from publicly available databases such as BRENDA.
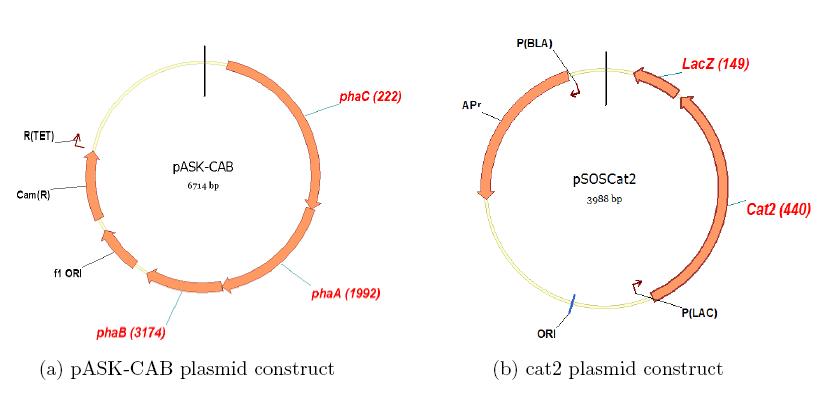
Similarly, we are currently attempting to construct a copolymeric circuit in E. coli.