Mississippi State/Problems
From 2007.igem.org
June 22, 2007: Ubiquitin-J01095 Construction
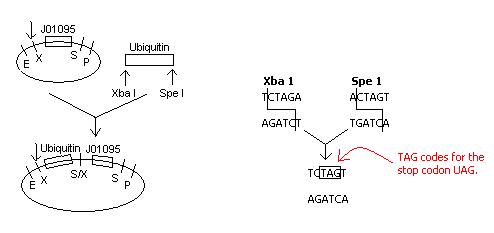
Upon inspection of our construction plans involving the insertion of ubiquitin next to the Registry part J01095, the S/X restriction site that would be created with the ligation of these two parts would code for the stop codon UAG.
Because it is not desirable for transcription to stop at the restriction site, a new plan of action must be developed.
Solution:
[http://www.abe.msstate.edu/wiki/msusbcwiki/index.php/#iGEM_2007:__Ubiquitin-Ligase_Probe
Plasmid Construction]
The right side is identical to that in the original construction. The left side will now include a fusion of E1010 (RFP) and ubiquitin, and primers will be customly designed and ordered specific to E1010 and ubiquitin. Instead of performing 2 transformations into 1 pET plasmid, we will co-transform 2 plasmids - one with GhR1-GFPuv fusion and one with RFP-ubiquitin - into the cell.
July 6, 2007: Low GhR1 Expression in Protein Gel
The results obtained from exposing the control (pGFPuv) and the GhR1-pGFPuv fusion to fluorescent light revealed that pGFPuv as a single part works as expected, as it shone brightly. However, the GhR1-pGFPuv showed no fluorescence, indicating that the amount of GhR1 was low, thus inhibiting the fluorescence of its fusion protein. Another factor contributing to low fluorescence of the fusion protein was that the cell strain used was XL1-Blue, a strain not noted for enhancing fluorescence.
Solution:
At this point in the project, it is necessary to consider changing the cell strain to one that would allow for stronger fluorescence (i.e. BL21(DE3)) and/or changing the plasmid. Currently, we are working with both XL1-Blue and BL21(DE3).
July 23, 2007: Unsuccessful Amplification of Ubiquitin
The first and second attempts at amplification of ubiquitin were unsuccessful. This could've been due to an incorrect assumption of the melting temperature; therefore, the lowest temperature reached during PCR would not have been low enough for the primer to anneal to the DNA. Another possible hindrance of amplification could've been the primer of choice. Also, the cDNA used for amplification was over 1 year old. Because cDNA is single-stranded, it degrades easily.
Solution:
The third PCR with the specific primer was successful, as Taq was used to amplify ubiquitin. Now, PFU will be used to amplify ubiquitin because PFU can proofread the newly synthesized sequence and correct possible mistakes. Additionally, reverse transcription PCR (1st strand cDNA synthesis) will be performed using cotton RNA. In another trial, the template for PCR will be ubiquitin itself, not cDNA as in all previous trials.
July 25, 2007: Lack of Protein Separation
The native protein gel used on July 24 contained 10% acrylamide/Bis, which prohibited the high molecular weight proteins of interest - GhR1-GFPuv fusion with and without ubiquitination - from separating much. Additionally, electrophoresis took place at 160 V for only 1 hour. Both of these factors contributed to vague results from the western blot, as the protein bands were very close together in the film.
Solution:
Native protein gel electrophoresis will be repeated using 8% acrylamide/Bis to allow the high molecular weight proteins to move downward more easily. Also, the gel will be run at 160 V for 2 hours. Increasing electrophoresis time and enhancing the separation of these proteins will provide for easier interpretation of them in the film after western blotting.
July 31, 2007: Incomplete Protein Expression
The GhR1 protein is in the order of 55 kDa, which is too large to be entirely expressed in soluble form. Additionally, misfolding is common among larger proteins.
Solution:
The GhR1 protein will be truncated before its conserved region, the Zn-finger domain. Since the Zn-finger domain is the only part of interest, another primer will be designed and ordered to allow amplification to begin at this conserved region. Shortening the protein will also reduce the chances of misfolding during amplification.
August 8, 2007: Restriction Site Within GhR1 Gene
There was an unexpected Eco R1 restriction site within the gene itself. This caused both PCR samples of GhR1 (one containing the conserved region only and the other containing the original, full-length gene) to be incorrect in sequence. This mistake was evidenced by the lanes of the agarose gel having two DNA bands per lane, instead of just one.
Solution:
The primer used to truncate the gene before its conserved region will be redesigned and ordered. The new primer will contain the restriction enzyme Spe 1 instead of Eco R1.
August 28, 2007: Incorrect GhR1 Sequence
When what was thought to be the GhR1 gene was analyzed to determine its sequence, it was discovered that the sequence was incorrect. Because the gene was amplified from cDNA, parts of the sequence were not consistent with our previously sequenced GhR1 gene.
Solution:
Earlier, the GhR1 gene had been ligated on the N-terminal of pGFPuv. This sequence was correct, so it will be used as a template to amplify the GhR1 gene. Also notable is that transformation will be done into BL21(DE3) (not XL1-Blue) since only a small amount of DNA is needed.