Vincent Parker Notebook
From 2007.igem.org
Vparker 17:11, 20 June 2007 (EDT) The sequences of the oligos I designed are as follows:
VP1AF-cgtaaAGATCTATGAGTCGTTTAGTCGTAGTATC VP2AIF-CGATGACATTATCTGGATtCACGATTATCACCTG VP3AIR-CAGGTGATAATCGTGaATCCAGATAATGTCATCG VP4AR-cgtatCTCGAGttaGGATCCTTACGCAAGCTTTGGAAAGGTAGC VP5BF-cgtaaAGATCTATGACAGAACCGTTAACCGA VP6BIF-GCAACGTATTACTCAGATtTGGCCACAAATGGCG VP7BIR-CGCCATTTGTGGCCAaATCTGAGTAATACGTTGC VP8BR-cgtatCTCGAGttaGGATCCTTAGATACTACGACTAAACG
The oligos were named according to my initials, numerical sequence, and letters determining direction of oligo. Ex. VP1AF- Vincent Parker 1 otsA Forward.
Vparker 15:04, 21 June 2007 (EDT)
After a long wait the oligos that I have ordered have arrived. I ordered 8 which PCR the two genes and get rid of the internal restriction sites. The template used to make the oligos was called MG 1655. In the ApE file which was split into two genes they were called ostA and otsB. They came in powder form and we have to dilute them down to 10 micro mols. That makes my master supply of my oligos when i use them i have to dilute them down to 100 micro mols. Using my oligos I can determine my PCR products... otsA will be broken up into two parts; PartA is 413 bps and PartB is 1077 bps. OtsB; PartA 485bps, PartB 381bps. I will PCR them overnight and digest tomorrow. Part A of otsA will be called V1-3 because i am using oligos 1 and 3 to PCr that specific part. Part B of otsA will be called V2-4. Part A of otsB will be called V5-7 and part B of otsB will be called V6-8. the two construction files look like this:
OTSA
PCR otsAF/otsAIR on otsA (413 bp, gp=A) PCR otsAIF/otsAR on otsA (1077 bp, gp=B)
PCR otsAF/otsAR on A+B (1456 bp BglII/XhoI) Digest pBca9145-Bca1144 (BglII/XhoI 2054+910, L) Product pBca9145-BBa I716301
otsAF forward BglII site anneals to otsA cgtaaAGATCTATGAGTCGTTTAGTCGTAGTATC otsAIF Removing BamHI site in otsA CGATGACATTATCTGGATtCACGATTATCACCTG otsAIR Removing BamHI site in otsA CAGGTGATAATCGTGaATCCAGATAATGTCATCG otsaR reverse XhoI site anneals to otsA cgtatCTCGAGttaGGATCCTTACGCAAGCTTTGGAAAGGTAGC
OTSB
PCR otsBF/otsBIR on otsB (485 bp, gp=A) PCR otsBIF/otsBR on otsB (381 bp, gp=B)
PCR otsBF/otsBR on A+B (832 bp, BglII/XhoI) Digest pBca9145-Bca1144 (BglII/XhoI, 2054+910, L) Product is pBca9145-BBa I716302
otsBF Forward BglII site anneals to otsB cgtaaAGATCTATGACAGAACCGTTAACCGA otsBIF Removes internal BglII site GCAACGTATTACTCAGATtTGGCCACAAATGGCG otsBIR Removes internal BglII site CGCCATTTGTGGCCAaATCTGAGTAATACGTTGC otsBR Reverse XhoI site anneals to otsB cgtatCTCGAGttaGGATCCTTAGATACTACGACTAAACG
VParker 16:37, 22 June 2007 (EDT) Big day planned. I have to take out my tubes V1-3, V2-4, V5-7, and V6-8 from the PCR machine.These are the 4 parts to my two genes. V1-3 is Part A of my otsA gene, it ends at the internal restriction BamHI site. V2-4 is part B to my otsA gene and it starts with the BamHI restriction site. I have already designed my oligos which code for a silent mutation within the BamHI site and codes for Leucine. V5-7 is part A of my otsB gene and ends at an internal BglII restriction site. V6-8 is part B of my otsB gene and starts at the BaglII restriction site and I have designed the oligos for that gene to have a silent mutation in the BglII site that codes for Isoleucine. As i came in to PCR my two genes, otsA and otsB, with the 8 oligos i designed. Two oligos per part. The oligos are named by number so when I mention V1-3 i am referring to part A of otsA but it will be PCRed with the first and 3rd oligos. Unfortunately when i came into the lab it was flooded and all I managed to do was take out Hannah's tube and my four out of the PCR machine and put them into the freezer for Monday. I did manage however to go home and sleep some more :-)
Vparker 16:02, 25 June 2007 (EDT)
Today i came into the lab and found that 3 out of the 4 PCR tubes were in the freezer but that i have lost the tube marked V6-8. I had the PCR products from the tubes V1-3, V2-4, and V5-7. After having fumed a bit over my lost PCR I went to work on setting up a new PCR tube for V6-8 and to also clean tubes V1-3, V2-4 and V5-7. Austin was kind enough to share the PCR machine with me and i put in the tube V6-8 in and also the sewing reaction i was working on named V1-4. V1-4 is a combination between V1-3 and V2-4. Since V1-3 and V2-4 are two parts of one gene and I had to PCR out the internal restriction site. 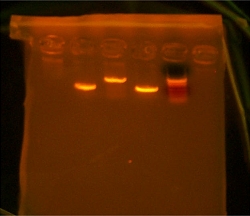 Decided to try my hand in Agar plates today so I volunteered the High School team to learn how to prepare and pour them since Arthur was not here. They turned out looking good with no bubbles. So i was behind today in terms of my PCR and i have to digest and ligate tomorrow instead of today. Losing the V6-8 tube set me back an hour and a half so i will save the rest for tomorrow.
Decided to try my hand in Agar plates today so I volunteered the High School team to learn how to prepare and pour them since Arthur was not here. They turned out looking good with no bubbles. So i was behind today in terms of my PCR and i have to digest and ligate tomorrow instead of today. Losing the V6-8 tube set me back an hour and a half so i will save the rest for tomorrow. 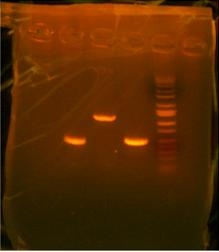
Vparker 15:04, 26 June 2007 (EDT)
Started off today with digestion of tubes V1-4 and V5-8 which are my genes that i PCRed. V1-4 was my otsA gene and V5-8 was my otsB gene. Atfer storing them in cultivation for an hour and a half Sam showed us how to prepare Agar solution and broth. we just put them in the autoclave machine and are waiting for both digestion and the autoclave to finish. There were problems with the autoclave because maybe the person who used it before us did not reset it after. Our Agar jars had to sit out while the machine exhausted itself and reset to take in our bottles. Nhu and I poured over 5 sleeves to help out the lab. The digestion of my tubes went well there were no problems and afterward I had to gel extract the DNA. I did not take a picture because too much exposure to UV rays is dangerous to my DNA and I wanted to keep as much of it as I could for the ligation process after I cut out the bright DNA bands from the gels. I had to melt the gels and put them through gel extraction recovery kit which binds the DNA to a filter and washes out all the enzymes and other solutions not wanted when I ligate the DNA to the vector. The number on the vector tube was 9145 and I named my ligation tubes otsA9145 and otsB1945. After ligation I had to heat shock the competant E.coli cells so that their membrane would become passive to the plasmid which contained my genes. I then used the little glass balls and smeared my competant cells with now hopefully my plasmid inside them. Tomorrow after I come in i will see if my bacteria grow. If not i am a complete failure :-(
Vparker 14:57, 27 June 2007 (EDT)
Came into work and had a nice spread of bacteria growing on my agar plates. On the agenda for today: colony PCR, which I am doing as I type this, followed by gel to make sure the colony I chose for PCR has the insert I want, and then I have to drop a colony into liquid media so that I can mini-prep for tomorrow. So far I am off to a rough start as I am only PCRing two colonies and I should have done at least 2 from each gene so that I can maximize my chances for success. I have basically done a gamble by only doing one colony for each gene and that chances of failure are high. With some great luck hopefully both were successful colonies and I won't have to re-PCR them. After lunch we will see after the gels if my colonies were chosen wisely. Just returning from lunch and PCR was already completed. Took 15 minutes to gel and the picture was perfect. The two genes I wanted to show within the bacteria are both present and my cloning process was a success. For today I have to transfer the bacteria to a liquid media and let incubate in a shaker over night. Tomorrow we will mini=prep the liquid media and then send off for sequencing. Also today we have a staff meeting at 3:00pm when I have to tell the group where I am and just an overall presentation of what I have done so far. Here is a picture of my gel form today. 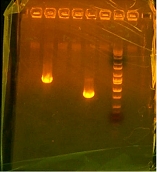
Vparker 16:37, 28 June 2007 (EDT)
After taking my 4 tubes out of the shaker, two tubes from each plate of colonies. All 4 of them where the proper color meaning that the plasmid was taken into the E.coli and the RFP was taken out. After mini-preping and accidentally drifting a little from the manual the DNA is in digestion right now to make sure the the plasmid that is in the Bacteria does in fact have our insert. I am digesting the plasmid with BglI and AlwnI which one cuts the insert and one cuts the plasmid. So if I have a band that does not correspond with what the digestion should be then that means the plasmid does not have my insert. If the bands are good then I can send them off for sequencing and tomorrow I will have to verify the sequences.