Dry to Wet
From 2007.igem.org
Contents |
Mass-Action Reaction Modelling
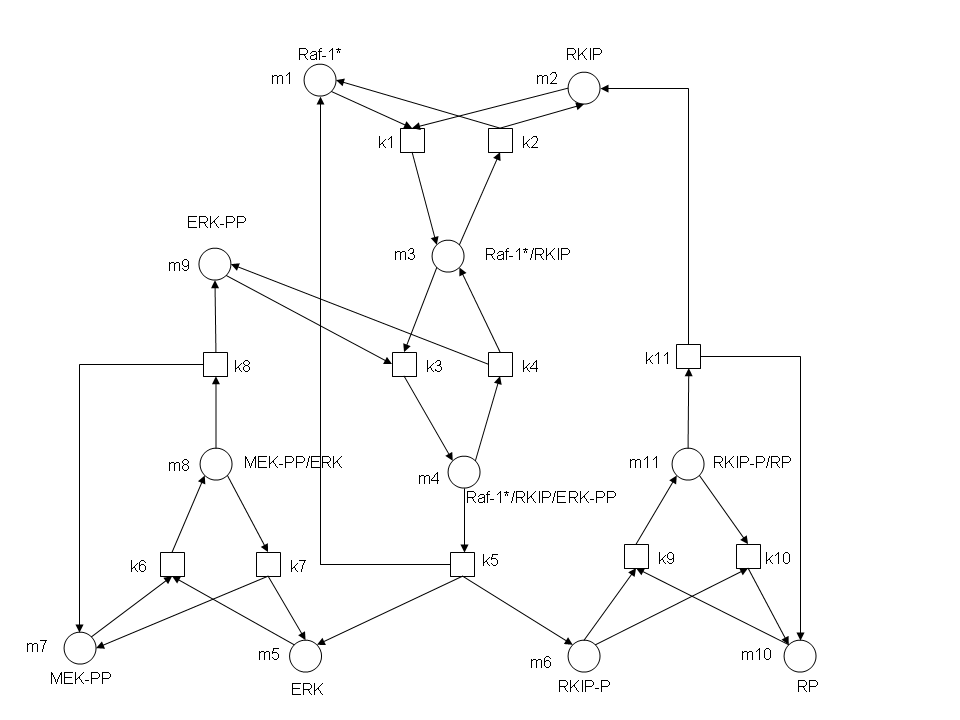
RKIP network
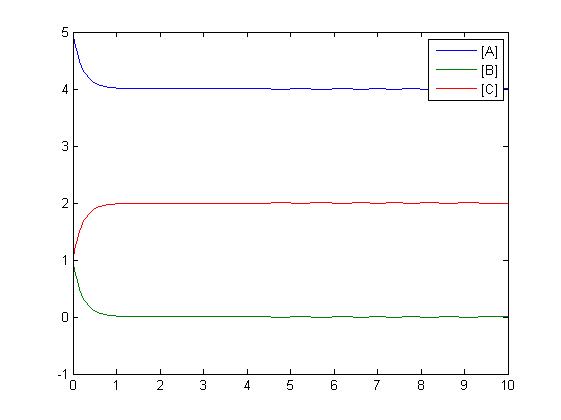
After gaining a thorough understanding of methods involved with modeling simple mass-action reactions, we can move on to more complex systems such as the RKIP network.
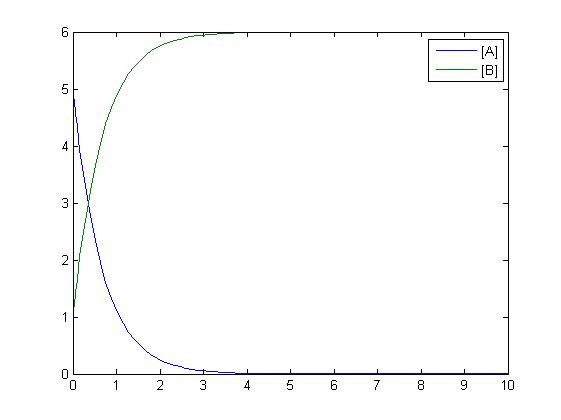
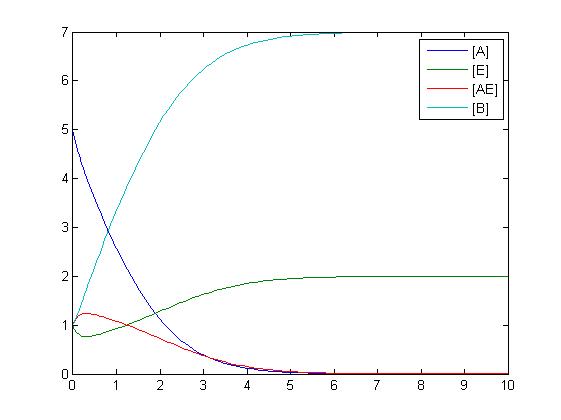
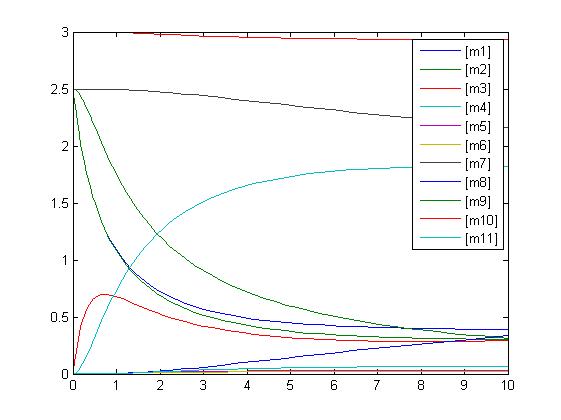
In the above diagram, substrates, enzymes and substrate/enzyme complexes are represented by numbered circles, rate constants are represented by numbered squares. By isolating individual species and their direct peripheral species (those being formed from or forming the isolated species) we are able to treat the group as a simple mass-action reaction. A differential equation is then found for each species based on the rate constants and code can be written and a graph plotted showing the trend of all the species’ concentration over time giving the following graph:
Sensitivity
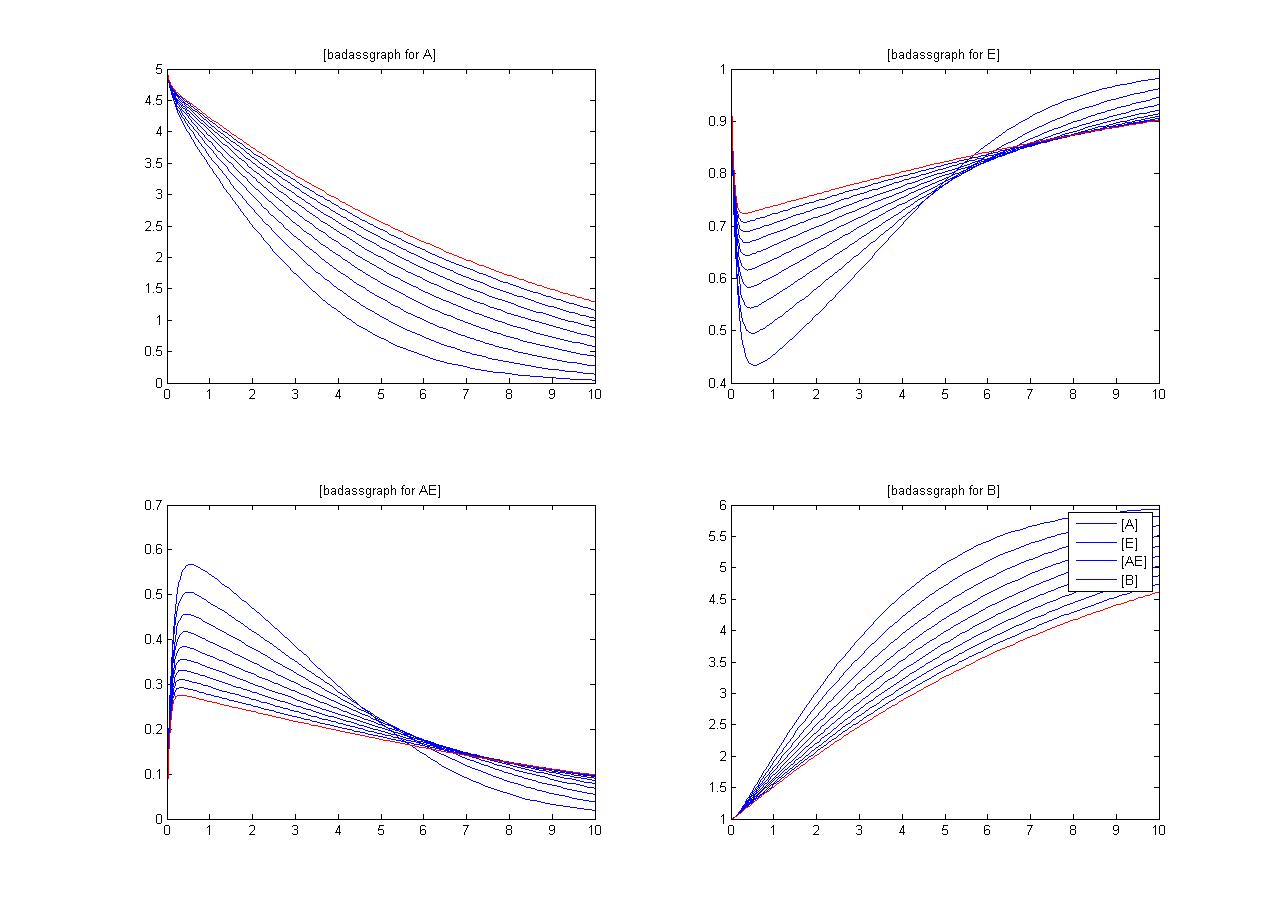
An insight into a system's sensitivity will show how the variation of a model can be apportioned qualitatively or quantitatively to different sources of variation
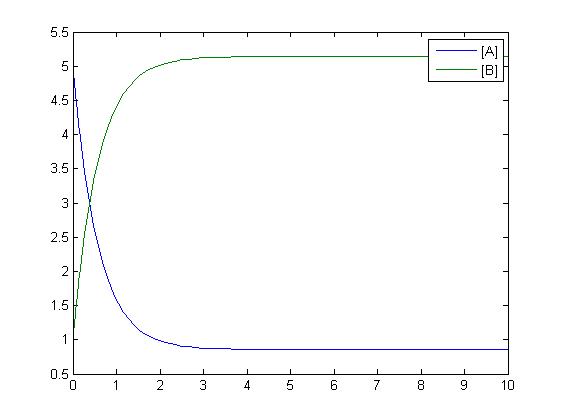
One method of exposing the variation of a model is to program a loop exposing a modelled reaction to increasing values of a chosen constant. This process was followed with the metabolic pathway showing in Mass-Action Reaction and ploted on a graph showing the response of all 4 species for a set range of varying K2 values from 1 to 10 where 10 is highlighted red.
Michaelis-Menton
Anybody who has done any sort of biological study will know michaelis menton what i am trying to acheive here is to take it from the basics so as to equate it to the equations we will be using to model the system and to give the biologists an idea of what values and models we need. The michaelis-Menton equation describes the kinetic properties of many enzymes. Consider the simple system A -> P The rate of V is the quantity of A that disappears over a specified unit of time which is equal to the rate of appearance of P. For this system V=k[A] where k is the rate constant. The simplest model that accounts for the kinetic properties of many enzymes is