BerkiGEM2007Present3
From 2007.igem.org
The Controller
The Controller is an integrated genetic circuit, comprised of two plasmids, that directs the initiation and production of the primary systems proteins through its component operons.
Introduction
The Bactoblood organism needs to exist in two different states: one form that is genetically stable and able to grow under normal laboratory conditions, and a second state that is highly differentiated, unable to grow, and devoid of genetic material. To bring about this transformation, we needed a controller that could be easily triggered to coordinate the differentiation process.
The controller required a large dynamic range between the off and on states, the ability to maintain and overexpress a large number of genes, and ideally employed a low-cost inducer. Therefore, we designed a controller based on a two plasmid system. One plasmid stably replicates the variou sbiosynthetic operons of our system at single copy in a transcriptionally-inactive state. The second plasmid houses the genes necessary for activation of the operon plasmid. When activated with iron, the copy number of operon plasmid increases to high-copy and the transcription of the operons is activated.
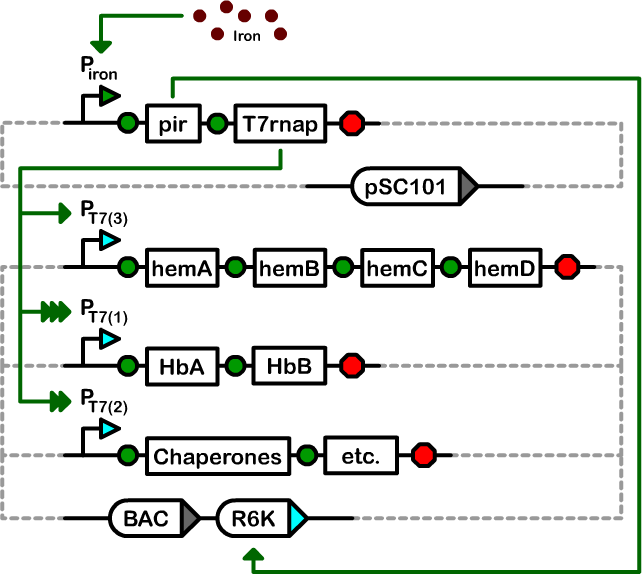
Our controller system is composed of two plasmids: a pSC101 Controller Plasmid, and an Amplifiable BAC.
Design and construction
We designed a two-plasmid architecture in which the biosynthetic operons reside on a single-copy bacterial artificial chromosome (BAC). The operons are under the transcriptional control of T7 promoters of various strengths. The BAC also contains an R6K origin of replication. In most strains of E. coli, this origin is silent as it requires the expression of the pir gene for replication. The second plasmid in our controller is a low-copy pSC101-derived plasmid that houses the T7 RNA polymerase and pir genes under the control of an iron-inducible promoter.
Construction of an iron-responsive PoPS-generating device
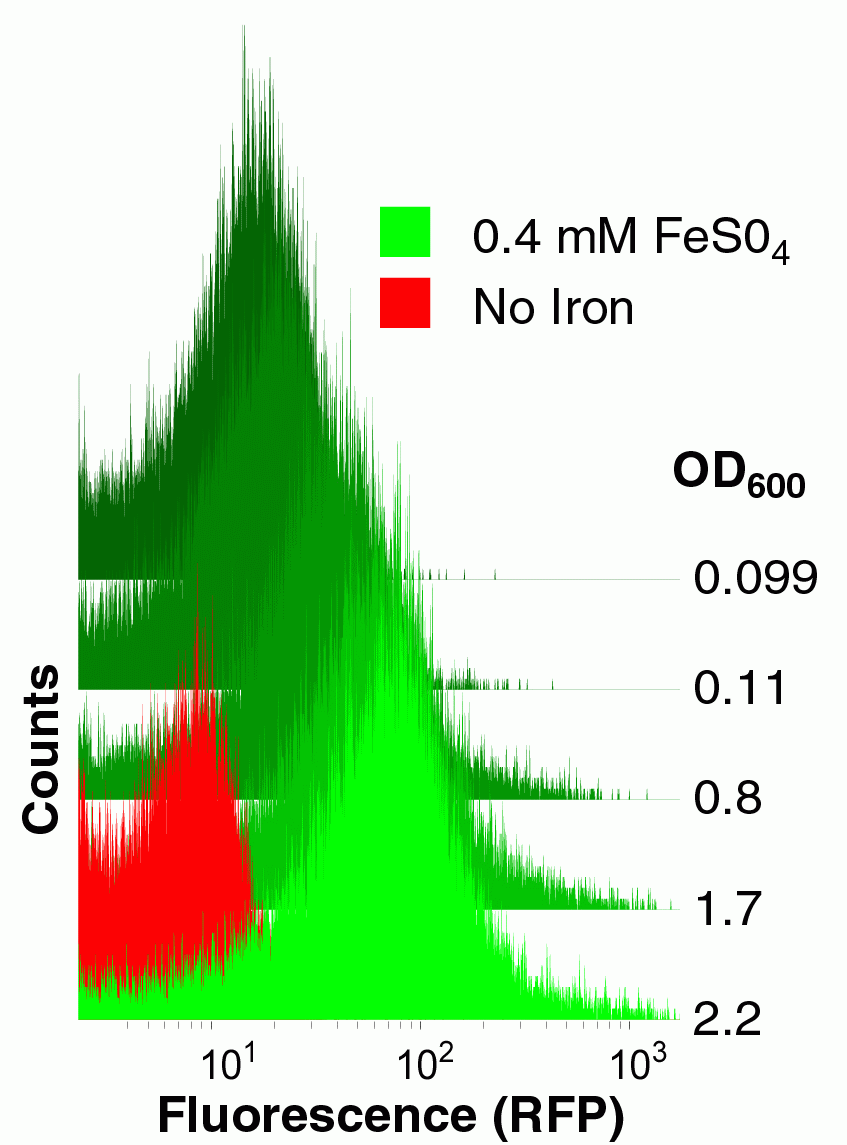
To construct this system, first we needed a promoter that was induced by iron. Microarray studies *ref* suggested that the yfbE promoter of E. coli might function as an iron-responsive PoPS-generating device. We therefore constructed a Biobrick derived from the yfbE promoter and constructed an RFP reporter composite part derived from this basic part. We examined the fluorescence of cells harboring this part both as a function of external iron concentration and growth phase. The yfbE promoter part had the ideal qualities for our controller: it is induced 100-fold as the bacteria emerge from the mid-log phase of growth, but only in the presence of exogenous iron.
Construction of an iron-dependent transcription device
To control gene expression with the controller, we needed to place the T7 RNA polymerase under the control of the yfbE promoter on a pSC101-derived plasmid. We therefore made a T7 RNA polymerase basic part and constructed a library of composite parts containing the yfbE part, one of nine ribosome binding site parts of different strengths, and the T7 RNA polymerase gene with a GTG or an ATG start codon. We constructed these composite parts in the pSC101 Biobrick plasmid I716101 and then examined their activity in an engineered E. coli strain, GH455G, containing a genome-integrated cassette with GFP under the control of a T7 promoter. Of the composite parts we constructed, only the composite part with the weakest ribosome binding site and a GTG start codon showed iron-dependent GFP production.
Construction of an iron-dependent copy number device
The third component of our controller was an iron-inducible copy number amplifier system. We therefore constructed a pir basic part and attempted to make composite parts with the yfbE promoter part. We were unable to construct Biobrick-derived cassettes of this composition apparently due to toxicity. Instead, we constructed a ribosome binding site-free version of the part in the pSC101-derived plasmid. We subsequently introduced a library of ribosome binding sites by EIPCR in which the start codon was either ATG or GTG and the ###-## positions relative to the ribosome binding site were randomized (See table ##). We introduced the library plasmids into E. coli cells harboring plasmid pBACr-AraGFP*ref* which is a BAC plasmid with an R6K origin of replication and GFP under an arabinose-inducible promoter. Due to the lack of a pir gene in these cells, only a low level of GFP production is observed when they are grown in arabinose media. In contrast, the production of GFP in
The pSC101 Controller Plasmid contains both the Self-Destruct Device, as well as the controller operon: The genes pir and the T7 RNA Polymerase under the control of an Iron Promoter. The pir gene activates the R6K origin, which causes the plasmid containing it to jump to high copy - the R6K origin is silent without pir. The T7 RNA Polymerase transcribes genes under the control of T7 Promoters, and will vary with strength depending on the variant of T7 Promoter. The T7 Promoters are silent without the T7 RNA Polymerase, and have a high level of transcription when active.
The Amplifiable BAC contains all the primary systems production genes, and two origins of replication. It contains both the BAC origin and the R6K origin, and so is single-copy when pir is absent, and high-copy when pir is active. Additionally, the BAC contains all the systems production genes, such as Hemoglobin, Heme (A to D), and the necessary chaperones, reductases, etc. Each of these operons is under the control of a custom T7 Promoter variant, tuned to produce the relative amount of protein desired for each operon. For example, the heme genes are under a weak T7 Promoter, as the heme genes naturally produce a large amount, where not too much is needed. However, the Hemoglobin operon is under a strong T7 Promoter, as we want a large amount of Hemoglobin present to effectively carry oxygen.