Brown
From 2007.igem.org
| Line 32: | Line 32: | ||
The architecture of our system is as follows: | The architecture of our system is as follows: | ||
[[Image:Tristable_Toggle_Switch_2007.jpg|100px|frame|left|The Tri-stable Toggle Switch Architecture]] | [[Image:Tristable_Toggle_Switch_2007.jpg|100px|frame|left|The Tri-stable Toggle Switch Architecture]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| Line 58: | Line 63: | ||
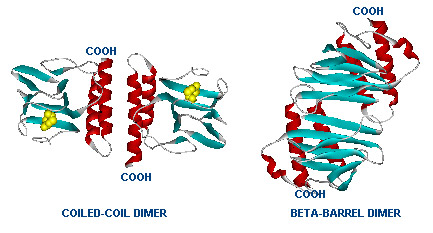
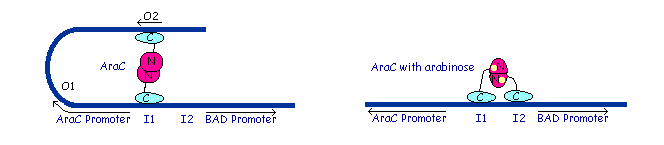
The gene AraC one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] The protein forms a dimer in with and without arabinose but the structural change activates or represses the pBAD ([http://en.wikipedia.org/wiki/Bcl-2-associated_death_promoter Bcl-2-associated death promoter], an apoptotic regulator in humans). | The gene AraC one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] The protein forms a dimer in with and without arabinose but the structural change activates or represses the pBAD ([http://en.wikipedia.org/wiki/Bcl-2-associated_death_promoter Bcl-2-associated death promoter], an apoptotic regulator in humans). | ||
| - | [[Image:Two_Dimers_of_AraC.jpg|Dimer structure with arabinose on the left (yellow)]] | + | [[Image:Two_Dimers_of_AraC.jpg|frame|Dimer structure with arabinose on the left (yellow)]] |
| - | [[Image:AraC_Promoters.gif|The left image shows the araC dimer repressing transcription, while the right conformation enables transcription]] | + | [[Image:AraC_Promoters.gif|frame|The left image shows the araC dimer repressing transcription, while the right conformation enables transcription]] |
====LacI==== | ====LacI==== | ||
====TetR==== | ====TetR==== | ||
Revision as of 19:14, 19 June 2007
Contents |
Welcome to our World
Projects
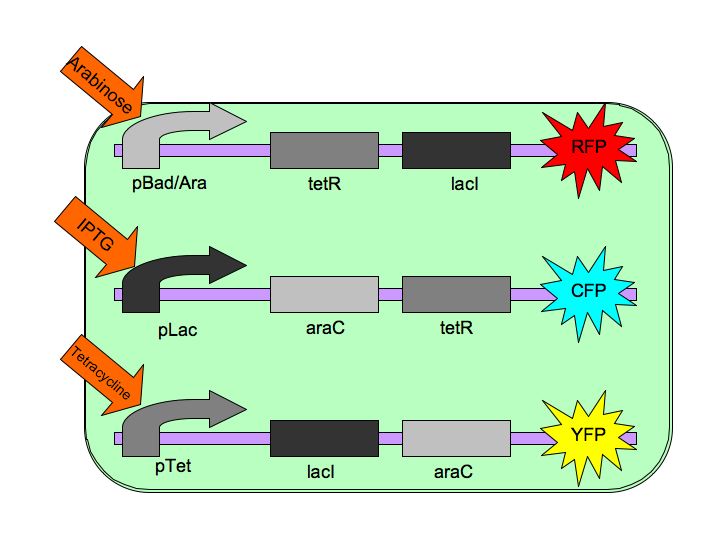
Tri-stable Toggle Switch
The Tri-stable Toggle Switch will be able to produce three distinct, continuous (stable) outputs for each of the three inputs. A chemical will induce the system to "lock into" one state while repressing the other two states. Our three constructs are pBAD->LacI->TetR, pLacI->AraC->TetR, and pTet->AraC->LacI, where AraC represses pBAD, LacI represses pLac and TetR represses pTet. The three chemicals ([http://en.wikipedia.org/wiki/Arabinose arabinose], [http://en.wikipedia.org/wiki/IPTG IPTG] and [http://en.wikipedia.org/wiki/Tetracycline Tetracycline], respectively), cause conformational changes in their respective repressor proteins which leads to gene expression. For example, in the presence of arabinose, AraC cannot repress pBAD so LacI and TetR are produced which in turn repress pTet and pLac.
The architecture of our system is as follows:
AraC/BAD
The gene AraC one of several genes (AraA, AraB, AraD, etc) originally for the metabolism of arabinose.[http://www.mun.ca/biochem/courses/3107/Topics/Ara_operon.html] The protein forms a dimer in with and without arabinose but the structural change activates or represses the pBAD ([http://en.wikipedia.org/wiki/Bcl-2-associated_death_promoter Bcl-2-associated death promoter], an apoptotic regulator in humans).