Chiba/Making Marimo
From 2007.igem.org
|
Introduction | Project Design ( 1.Affinity Tag | 2.Communication Module | 3.Size Control ) | Making Marimos | Our Goal |
Making Marimos
Parts Construction
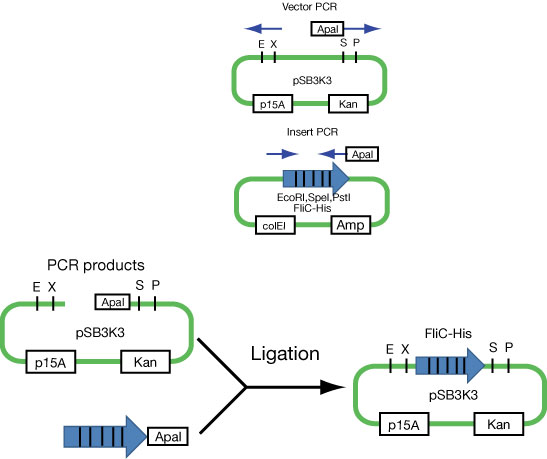
Because we found that FliC code includes restriction enzyme(EcoRI,SpeI,PstI) which is used in Biobrick, we divided into plasmid of FliC and one of signal, and aimed the double transformation.
Moving FliC-His generator
Experiment
- We regulate the expression of His-tagged FliC by lux promoter. Namely, if LuxR is expressed, bacteria can express FliC via Quorum Seinsing.
- Because the Quorum Sensing device is on ColE1-type vector, we need to export the FliC unit into the plasmids with compatible origins such as p15A.
Results
First attempt to import FliC-His generator into p15A vector (see Fig).
- Communication units are on ColE1 type plasmids. To make this stickly FliC construct compatible with communication circuits, this is an absolute necessity.....
- Many forbidden sites for Biobrick production throughout the FliC gene. We are planning to eliminate them all in near future by repeated site-directed mutagenesis using ExSite method.
FliC-his biobrick
Experiment
- Making Biobrick version of Flic-His and other stickly-FliCs. unfortunately, FliC has many EcoRI,SpeI,PstI sites. We have to eliminate them one by one by site-directed mutagenesis.
Results
Not yet (as of 10/26/2007). The first ligation sucked.
Cloning is still underway