Christopher Anderson Notebook
From 2007.igem.org
JCAnderson (Talk | contribs) (→JCAnderson 11:56, 9 September 2007 (EDT)) |
JCAnderson (Talk | contribs) |
||
| Line 1: | Line 1: | ||
==~~!~~== | ==~~!~~== | ||
| - | ==[[User:JCAnderson|JCAnderson]] 21: | + | ==[[User:JCAnderson|JCAnderson]] 21:32, 11 September 2007 (EDT)== |
| - | I | + | [[Image:BerkiGEM2007_JCA091107image1.jpg|left|75px]] |
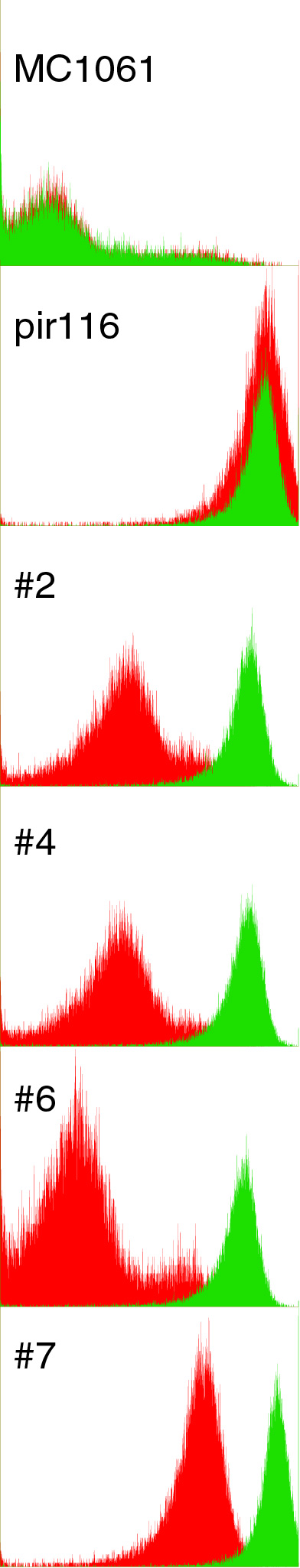
| + | I did 1mL cultures in LB+100ug/mL ara of the pir controllers of pBACr-AraGFP with and without 5mM iron. I grew for 5 hrs at 37 in a block, pelleted, then did cytometry. Red is no iron, green is with iron. Looks mighty nice! I've decided to examine 2, 6, and 7 further, so I mini'd, diluted 10x, and transformed lefty cells to sep. | ||
==[[User:JCAnderson|JCAnderson]] 11:56, 9 September 2007 (EDT)== | ==[[User:JCAnderson|JCAnderson]] 11:56, 9 September 2007 (EDT)== | ||
[[Image:BerkiGEM2007JCA090907image1.jpg|300px|left]] | [[Image:BerkiGEM2007JCA090907image1.jpg|300px|left]] | ||
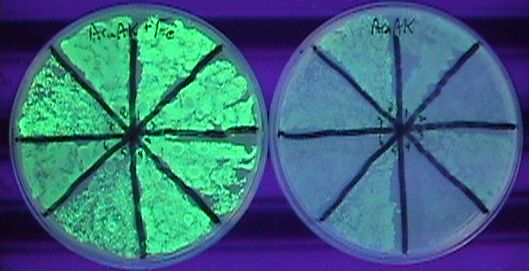
Alright, that took a while. What ended up working was taking the yfbE-pir composite part (no rbs), dropping that in the pSC101 plasmid, then doing EIPCR with dt013/14, and transforming a whole lot of material by electroporation. I only had about 50 members to the library in the end, but there were plenty of hits in there. I'm not sure how many, because transforming the lib with pBACr-AraGFP came out severely contaminated. I was able to find green colonies, though, and picked 8 of them, plating with and without iron (and Ara to turn on GFP). Several of those look nice and tight. | Alright, that took a while. What ended up working was taking the yfbE-pir composite part (no rbs), dropping that in the pSC101 plasmid, then doing EIPCR with dt013/14, and transforming a whole lot of material by electroporation. I only had about 50 members to the library in the end, but there were plenty of hits in there. I'm not sure how many, because transforming the lib with pBACr-AraGFP came out severely contaminated. I was able to find green colonies, though, and picked 8 of them, plating with and without iron (and Ara to turn on GFP). Several of those look nice and tight. | ||
| + | |||
| + | ==[[User:JCAnderson|JCAnderson]] 21:35, 20 August 2007 (EDT)== | ||
| + | I'm going to finish off the iron-pir construct David started. I set up a DT013/DT014 EIPCR on I716119 Clone 1 with 4K55 Expand. I716119 is the promoter-pir construct with no rbs in pBca9145. The oligo will put in an rbs library. i'll sub with SpeI/DpnI tomorrow. | ||
Revision as of 01:32, 12 September 2007
Contents |
~~!~~
JCAnderson 21:32, 11 September 2007 (EDT)
I did 1mL cultures in LB+100ug/mL ara of the pir controllers of pBACr-AraGFP with and without 5mM iron. I grew for 5 hrs at 37 in a block, pelleted, then did cytometry. Red is no iron, green is with iron. Looks mighty nice! I've decided to examine 2, 6, and 7 further, so I mini'd, diluted 10x, and transformed lefty cells to sep.
JCAnderson 11:56, 9 September 2007 (EDT)
Alright, that took a while. What ended up working was taking the yfbE-pir composite part (no rbs), dropping that in the pSC101 plasmid, then doing EIPCR with dt013/14, and transforming a whole lot of material by electroporation. I only had about 50 members to the library in the end, but there were plenty of hits in there. I'm not sure how many, because transforming the lib with pBACr-AraGFP came out severely contaminated. I was able to find green colonies, though, and picked 8 of them, plating with and without iron (and Ara to turn on GFP). Several of those look nice and tight.
JCAnderson 21:35, 20 August 2007 (EDT)
I'm going to finish off the iron-pir construct David started. I set up a DT013/DT014 EIPCR on I716119 Clone 1 with 4K55 Expand. I716119 is the promoter-pir construct with no rbs in pBca9145. The oligo will put in an rbs library. i'll sub with SpeI/DpnI tomorrow.