David Tulga Notebook
From 2007.igem.org
(→Bca1106-H RBS + T7rnap GTG (I716117) - Building (Growing in Culture)) |
(→Bca1106-H RBS + T7rnap GTG (I716117) - Building (Growing in Culture)) |
||
| Line 218: | Line 218: | ||
The sequencing results came back all as some variation on a truncation recombination, indicating that I will need to re-make the 116 library. | The sequencing results came back all as some variation on a truncation recombination, indicating that I will need to re-make the 116 library. | ||
| - | ==Bca1106-H RBS + T7rnap GTG (I716117) - | + | ==Bca1106-H RBS + T7rnap GTG (I716117) - Confirmed== |
'''Construction File:''' [[BerkiGEM2007-DavidConstructionFile117 | I716117: Bca1106-H RBS + T7rnap GTG]] | '''Construction File:''' [[BerkiGEM2007-DavidConstructionFile117 | I716117: Bca1106-H RBS + T7rnap GTG]] | ||
Revision as of 00:17, 8 August 2007
<< Back to UC Berkeley iGEM 2007
My Construction Files
My Sequencing Files
Notebook Archives: May 30 - June 5, Finished Basic Parts 101-107
Progress Status Descriptions
Designing : Making Construction Files and Ordering Oligos
Building : PCR, Transforming, Culturing
Checking : Analytical Digesting and Sequencing
Confirmed : Correct clones are confirmed and mini/midiprep'd
Finished : Correct clones are organized and saved
Composite Part Construction
RBS Library + pir (I716108) - Finished
Construction File: I716108: RBS Library + pir
June 27: I digested the pir confirmed part (clone #4) with EcoRI and BglII, then ligated this to the digestion of pBca1122-Bca9235 with EcoRI and BamHI. I then transformed these into some DH10B cells, and plated on AMP+KAN plates today, and will grow the culture tomorrow.
June 29: Due to the incubator being turned off, I could not grow the culture yesterday, as the colonies were not visible, therefore, I scraped the plate in LB today, and grew in AMP+KAN LB Media (which I had to make, due to our being out of that particular mix), to then miniprep on Monday. (After being placed in the refrigerator on Saturday)
July 2: I have miniprep'd the library, and it appears correct, but I will not know whether the RBS + pir combination works until I test it with a promoter in the next pir composite part.
July 25: The sequencing result of some individual clones of 109 came back, indicating the RBS library must be correct.
July 27: I have now saved this library on the clone saver sheet.
Salicylate Promoter + RBS Library + pir (I716109) - Finished
Construction File: I716109: Salicylate Promoter + RBS Library + pir
July 2: I digested the RBS+pir (108) part with EcoRI and BglII, Gel Purifying the large fragment. As well, I digested the Salicylate Promoter with EcoRI and BamHI, Gel Purifying the small fragment, for an insertion. I then ligated the two fragments together, and transformed, and plated on an AMP plate, as the KAN resistance gene should have been excised. However, the digest of the RBS+pir library looked very smeary, as there was likely incomplete digestion.
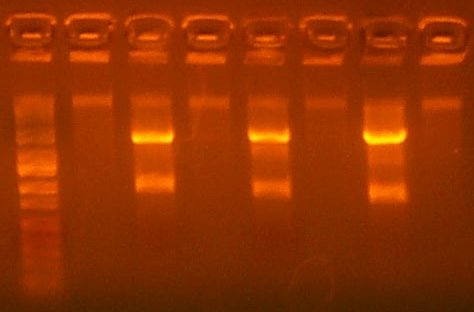
| The Test Digest Gel. Lane 1 is marker, lanes 2-9 are the T7rnap test Digests, the second to last is the RBS+pir library digest, and the last lane is the Salicylate Promoter digest. |
July 3: I checked the plate, but no colonies grew, I think this is likely due to the incomplete digestion that I observed yesterday, and there being an insufficient amount of ligated product to transform with.
July 4: I again digested the RBS+pir (108) part with EcoRI and BglII, Gel Purifying the large fragment, with BglII and XhoI, Gel Purifying the small fragment, as well as digesting the Salicylate Promoter with BamHI and XhoI, Gel Purifying the large fragment, with all digests this time for two hours, to ensure complete digestion.
July 5: I have ligated the EcoRI fragements together as well as the XhoI fragments together, to hopefully ensure at least one insertion will work. I then transformed and plated on AMP plates, as the KAN resistance should have been excised out. I will then scrape and grow in AMP media tomorrow.
July 6: Neither of the plates looked sufficient, with only a few colonies. I believe the ligase I was using might be bad, so I have attempted the same ligation/transformation/plating on AMP as yesterday, but this time with a different ligase.
July 9: Again, the plates looked very insufficient to no colonies, so I will have to re-do the transformations over again, possibly with a different procedure.
July 10: I have re-done the same ligation / transformations with 4 times as much miniprep digest, and in TG1 cells, to hopefully to get more colonies.
July 11: The plates looked a little better, but still very low yield. I have tried the EcoRI digests, etc. again, as it was the most successful direction in the previous attempts.
July 12: The plate looked excellent, and so I scraped and cultured it in 5mL of LB+AMP Broth.
July 13: I have miniprep'd the library, and digested it and the 101 low copy plasmid with BglII and XhoI. 101's large fragment (backbone) was gel purified, but 109's bands were too close to differentiate, so I ran a ca998/G00101 PCR on it to amplify the part.
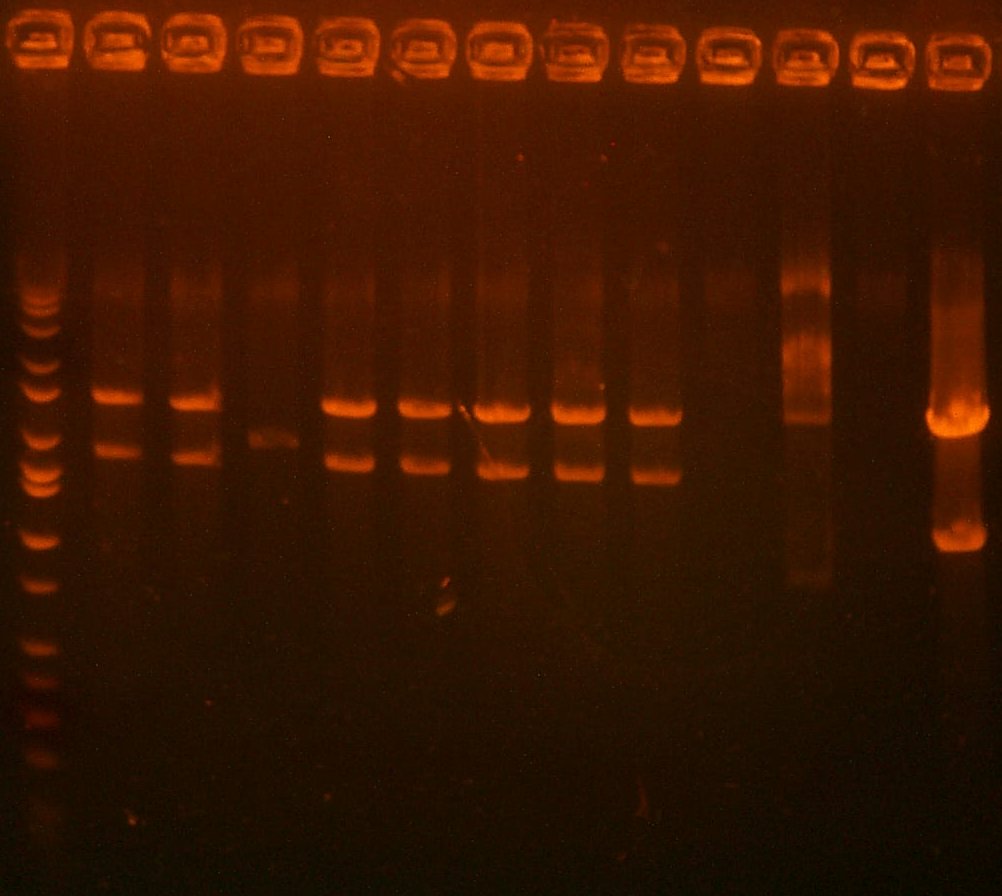
| The Digest Gel. Lane 1 is the 109 digest, lane 2 is the 101 digest, and the last lane is the Marker. |
July 16: I gel-purified the PCR Product, and while there were multiple bands, the desired one was considerably brighter than the rest. I then digested the PCR Product with BglII and XhoI. I then transformed the ligation of 109 in 101 into pBACr-AraGFP cells, and plated on an AMP+KAN+Ara+Sal plate.
July 17: The 109 in 101 ligation product had lots of colonies, but no obviously green ones, so I picked 96 and grew in 600 uL of AMP + KAN + Sal + Ara.
July 18: All 96 wells showed uniform uninduced activity, even with Salicylate in the media, so I miniprep'd 2 of them and did a PCR with ca998 / pir-R, for use in sequencing tomorrow.
July 19: I did another digest of 101 with BglII and XhoI, in case my previous digest was too low-quality. Additionally, the PCR for the two wells failed, so I will have to sequence a high copy plasmid. I also transfomed a new ligation of 109 in 101 on AMP+KAN+ARA+SAL, as well as directly transformed 109 on that kind of plate. I also transformed the ligation product of only the digest itself, to test the quality of the digests.
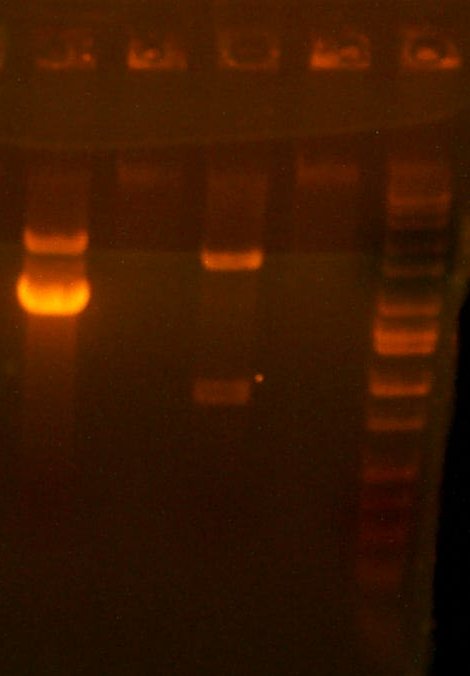
| The Digest Gel. Lane 1 is Marker, 2 is the 101 digest, and lane 3 is a T7rnap EcoRI/BglII digest. |
July 20: The first 101 digest looked clean, the second had many colonies, and the 109 digest had lots of colonies. Additionally, the 109 direct and 109 in 101 plates showed no significant activity. Therefore, I transformed into TG1 cells 1 uL of a 20x dilution of 109 onto AMP to then pick and analyze individual colonies. I also test digested with HindIII the 109 library to see if its any good. The band pattern looked approximately right, but somewhat fuzzy and uncertain.
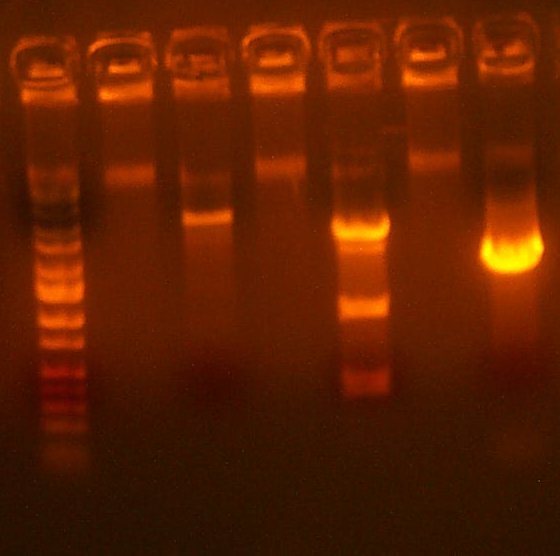
| The Digest Gel. Lane 1 is Marker, 2 is the 101 digest, lane 3 is the 109 test digest with HindIII, and the last lane is the PCR for the low/high plasmid Arthur is making. |
July 23: I picked 4 colonies of 109 to then miniprep and sequence, to determine if the construct is correct.
July 24: I have miniprep'd the 4 109 colonies, and sent off for sequencing with ca1111F, which binds to the middle of the nahR gene in the Salicylate promoter part.
July 25: The 4 clones came back as correct, and so this library is correct, even though it does not appear to work, most likely due to the ineffectiveness of the Salicylate promoter.
July 27: I have now saved this library on the clone saver sheet.
RBS Library + T7rnap ATG, GTG (I716110, I716111) - Confirmed
Construction File: I716110,I716111: RBS Library + T7rnap ATG, GTG
July 4: I digested the T7rnap ATG and GTG (103,104) parts with EcoRI and BglII, Gel Purifying the large fragment. I performed this digest for 2 hours, as previous digests were somewhat smeary.
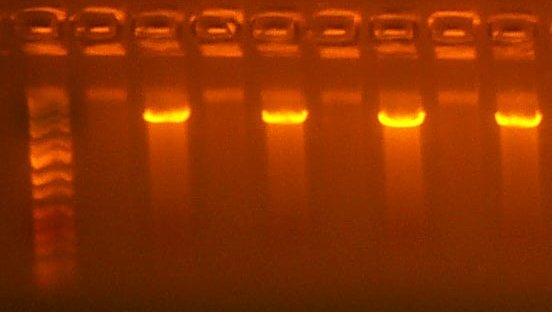
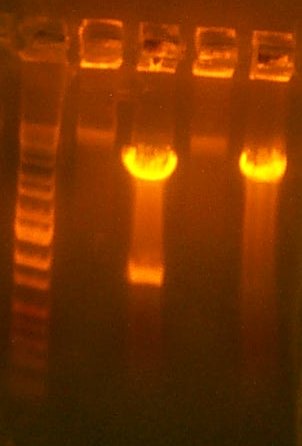
| The Digest Gel. Lane 1 is marker, All digests are EcoRI and BglII, and the clone order is: 103 #2, 103 #4, 104 #2, 104 #3. |
July 5: I ligated the EcoRI/BglII digest with the EcoRI/BamHI digest of the RBS Library, and then transformed some DH10Bs and plated on AMP Plates, to then grow in AMP/KAN Media tomorrow.
July 6: None of the plates looked sufficient, with none to a few colonies. I believe the ligase I was using might be bad, so I have attempted the same ligation/transformation/plating on AMP+KAN as yesterday, but this time with a different ligase.
July 9: Again, the plates looked very insufficient to no colonies, so I will have to re-do the transformations over again, possibly with a different procedure.
July 16: I have digested the new T7rnap parts with EcoRI and BglII, to then insert the RBS part. I have ligated in the RBS part, and transformed into TG1 cells, and plated on AMP+CAM.
July 17: I have scraped and cultured the plates in AMP+CAM, as there were a ridiculous number of colonies.
July 18: I have now miniprep'd the two libraries, as they appear to be correct.
July 26: As the phenotypic analysis indicates definite functionality in both high and low copy plasmids, this library is confirmed to be correct.
Salicylate Promoter, Iron Promoter + RBS Library + T7rnap ATG, GTG
(Salicylate: I716112, I716113; Iron: I716114, I716115) - Confirmed
Construction Files: I716112,I716113: Salicylate Promoter + RBS Library + T7rnap ATG, GTG
I716114,I716115: Iron Promoter + RBS Library + T7rnap ATG, GTG
July 19: I have digested the T7rnap ATG and GTG RBS Libraries (110,111) with EcoRI and BglII and purified the Large fragment. This was then ligated with an EcoRI and BamHI digest of the Salicylate Promoter and Iron Promoters, and transformed into some TG1 cells, and plated on AMP.
July 20: The T7 GTG Starts have a decent colony yield, but the ATGs don't have very many colonies. This is likely due to the toxicity of the T7rnap, as there is some leaky expression even when uninduced. Therefore, I have scraped and miniprep'd directly off the plate for the GTG starts. I also re-did the ATG (110) Ligation / Transformation with more T7rnap material into TG1s and plated on AMP.
July 23: The T7rnap ATG plates have a sufficient colony yield, but not too many, indicating high toxicity. I then scraped and miniprep'd the Promoter/RBS/T7RA plates. I have also digested the Promoter/RBS/T7RA,G plasmids with BglII and XhoI and ligated into a BglII/XhoI digest of 101, to make them low copy. I then transformed into GH455G cells on AMP: The Ligations of 112-115 in 101, and the direct plasmids of 112-115, and 112-113 on AMP+SAL.
July 24: These is confirmed activity with the Iron Promoter with the ATG and GTG Starts, and the low copy ATG starts have very few colonies. I have then picked 96 colonies on a 96-well plate from the low copy GTG starts - columns 1-8 are 115 (Iron) in 101, columns 9-12 are 113 (Salicylate) in 101, and put this in the incubator to grow overnight, with TB+AMP+Glycerol.
July 25: I grew up the 96 well plate (10 uL of culture from each well) in two 96 well blocks (600 uL culture in each well), one with LB+AMP only, and the other with LB+AMP+Iron or Salicylate, depending on the construct, to then do TECAN analysis when the cultures are at saturation.
July 26: I have done the TECAN analysis with 10uL of each well (on/off), with the parameters of bandwidth = 7.0, GFPmut2 (Ex: 481nm, Em: 507nm) as the spectrum, and a gain of 80. The well D4 was a hit with a variance of 1850 (off: 1300, on: 3150), or 4.3 times a fluorescent, after removing the background. I have also miniprep'd the D4 well (off), and set up a ca998/T7-R PCR to amplify the part for sequencing.
July 27: The D4 PCR failed, so I ran an Iron-F/T7-R PCR on D4 to then sequence.
July 30: The D4 PCR was successful this time, so I gel purified it, and sent it off for sequencing with the ca570F oligo, which is a reverse internal T7 sequencing oligo.
July 31: The sequencing result came back correct, with the 1106-H RBS.
Iron Promoter + RBS Library + pir (I716116) - Building (Re-Transforming)
Construction File: I716116: Iron Promoter + RBS Library + pir
July 25: I have ligated the Iron Promoter digest with EcoRI and BamHI, Small Part Fragment, and the EcoRI/BglII, Large part fragment, digest of 108 (RBS+pir). I then transformed in TG1s, and plated on AMP.
July 26: I have now scraped and cultured the 116 (Iron+108) library, as the colony yield was very good.
July 27: I have miniprep'd the 116 (Iron+108) library, and digested it with BglII and XhoI, as well as digested 101 with BglII and XhoI. I then ligated the digests of 116 and 101, transformed in TG1, plated on AMP, to then use these for the EIPCR Quickchange oligos to create a larger library of RBS's. I also transformed 116 directly in pBACr-AraGFP and plated on AMP+KAN+ARA, to test if there was any obvious activity in high copy.
July 30: There is no obvious activity in high copy, and the 116(101) plate had a reasonable colony yield, and so I picked 4 colonies to grow in AMP and sequence.
July 31: All the cultures were red in the pellet except for #2, indicating they were low-copy plasmid bleedthrough. I then miniprep'd #2 and sent it off for sequencing.
Aug 1: The sequencing came back as a high-copy Truncation Recombination, and so it is incorrect. I have picked and grown in AMP 4 more 116(101) colonies, that do not appear red, to miniprep/test/sequence. I have also tried an extra-long gel purify of the 116 and 101 digests, to clean up the bleedthrough. I then ligated this and transformed into DH10Bs, and plated on AMP. I also did a ligation digest check plating the 116 and 101 digests in DH10Bs on AMP. I also transformed a dilution of the 116 library into DH10Bs on AMP to pick a few to then use a single clone to insert into the low-copy plasmid. I also transformed some DH10Bs on AMP with 101 to then make more with a midiprep.
Aug 2: The 116(101) transformation failed, and there are very few/no colonies on the 101 and 116 digests. I also miniprep'd the 116(101) cultures I picked yesterday from the first transformation, and ran 8ul on a gel. All the bands are bright and are high-copy bleed-through. I have also picked a few of the 116 colonies and grew in AMP, to then miniprep and sequence. Finally, I also picked a 101 colony to make more with a midiprep.
Aug 3: I performed a midiprep of the 101 plasmid, and the pellet appeared slightly red, indicating it is correct. I also miniprep'd the 116 cultures, and sent off for sequencing.
Aug 4: The sequencing results came back all as some variation on a truncation recombination, indicating that I will need to re-make the 116 library.
Bca1106-H RBS + T7rnap GTG (I716117) - Confirmed
Construction File: I716117: Bca1106-H RBS + T7rnap GTG
July 31: I have digested the 1106-H RBS with EcoRI and BglII (L), and the 1122 plasmid with EcoRI and BglII (S), to add the KAN resistance gene to the RBS. I then ligated these two fragments and transformed into TG1s on AMP+KAN.
Aug 1: The colony yield for the KAN+RBS was excellent, so I picked 4 colonies and grew in AMP+KAN to miniprep.
Aug 2: I have miniprep'd the 4 KAN+RBS clones, and test digested them with EcoRI/BamHI. All clones look correct, and so I have cut out and gel purified the Small fragment (1047) of 2 of them. I then ligated this fragment (1) with the T7RG (104,1) EcoRI/BglII Digest, and transformed into DH10Bs on AMP+KAN.
Aug 3: The colony yield for the KAN+RBS+T7RG was very good, so I picked 4 clones and grew up in AMP+KAN, to then miniprep and sequence.
Aug 6: I have miniprep'd the RBS+T7RG (117) clones, and sent off for sequencing. A quick test gel indicates that the sizes are good, indicating they are not RBS-only bleedthrough.
Aug 7: The sequencing came back, and all 4 clones are perfect.
Strain and Competent Cell Production
T7GFP Strain - GH455G
July 2: Chris took the -80 of this strain out of the freezer, and plated it on an LB-Only plate.
July 3: I picked a colony from the plate, and grew it up in 5mL of LB-Only Broth.
July 4: I took the culture out of the incubator, and put it in the refrigerator, to enable simultaneous competent cell production with the pBACr-AraGFP Strain.
July 5: I made competent cells from the culture grown on the 3rd, for a total of approximately 25 200uL aliquots. I then froze them using liquid nitrogen, and placed them in a box in the top shelf of the -80 freezer.
July 6: I then transformed some of the competent cells with pBAC-T7621B, as this plasmid produces the T7rnap constitutively, and should indicate whether the cells are indeed competent and functional. These were then plated on a KAN-only plate.
July 9: The transformation with pBAC-T7621B worked very well, and the transformed cells do, in fact, glow green under UV light, indicating that the cells are indeed competent, and functional.
pBACr-AraGFP (in DH10B) Competent Cells
July 2: I have transformed some DH10B cells with pBACr-AraGFP, and plated on a KAN-only plate.
July 3: Yesterday's transformation failed, as there were no colonies. Therefore, I have tried transforming some DH10B cells with pBACr-AraGFP again, this time with a longer rescue (>1 hour, in 2yT), and plated them on a KAN-only plate.
July 4: The plate appeared to have very few colonies, but they did have the resistance gene, so I picked a colony from the plate, and grew it up in 5mL of LB+KAN Broth.
July 5: I made competent cells from the culture grown yesterday, for a total of approximately 25 200uL aliquots. I then froze them using liquid nitrogen, and placed them in a box in the top shelf of the -80 freezer.
July 9: I transformed some of these competent cells with I716107 to give them AMP resistance, and have plated on an AMP + Arabinose plate, to test their competency and functionality, as they should grow slightly green upon induction with Arabinose.
July 10: The plate I grew yesterday has hundreds of colonies, and glows slightly green under UV light, indicating that the cells are indeed both competent and functional.
July 17: I am testing the cells' sensitivity to AMP, by plating directly some competent cells on an AMP plate.
July 18: The cells are indeed AMP-sensitive, as the AMP-only plate appears clean, with no colonies whatsoever.