Davidson Missouri W/Results
From 2007.igem.org
Wideloache (Talk | contribs) (→Part Flipping and Computation) |
(→Results) |
||
| Line 16: | Line 16: | ||
In addition to the regular split GFP and split RFP constructs, we also wanted to test some "hybrid" constructs to ensure that they would not fluoresce. GFP and RFP have similar structures and functions. Previous studies have shown that it is possible to modify GFP to display a wide range of color phenotypes. We created the following parts: Plac-RBS-RFP1-hixC-GFP2 and Plac-RBS-GFP1-hixC-RFP2. A plasmid may, at some point during its flipping process, contain such sequences. We tested to make sure that the similarity of these two proteins did not make them compatible enough to fluoresce. It was found that neither of these parts show any fluorescence or color change. | In addition to the regular split GFP and split RFP constructs, we also wanted to test some "hybrid" constructs to ensure that they would not fluoresce. GFP and RFP have similar structures and functions. Previous studies have shown that it is possible to modify GFP to display a wide range of color phenotypes. We created the following parts: Plac-RBS-RFP1-hixC-GFP2 and Plac-RBS-GFP1-hixC-RFP2. A plasmid may, at some point during its flipping process, contain such sequences. We tested to make sure that the similarity of these two proteins did not make them compatible enough to fluoresce. It was found that neither of these parts show any fluorescence or color change. | ||
| + | |||
| + | ==Constructs== | ||
| + | |||
| + | Once we managed to split two genes we proceeded to implement two different graphs in plasmids. | ||
| + | |||
| + | We built the following constructs: | ||
| + | *Graph A <br>[[Image:hpp-1.png]] | ||
| + | *Graph B <br>[[Image:hpp-2.png]] | ||
| + | *Solution to both A and B <br>[[Image:hpp-3.png]] | ||
| + | *Positive Control 1 | ||
| + | *Positive Control 2 | ||
==Part Flipping and Computation== | ==Part Flipping and Computation== | ||
Revision as of 19:08, 23 October 2007
Contents |
Results
Gene Splitting
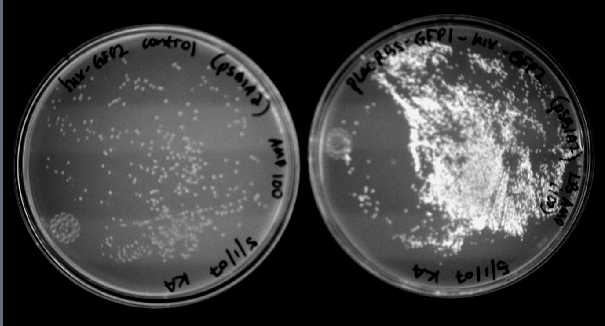
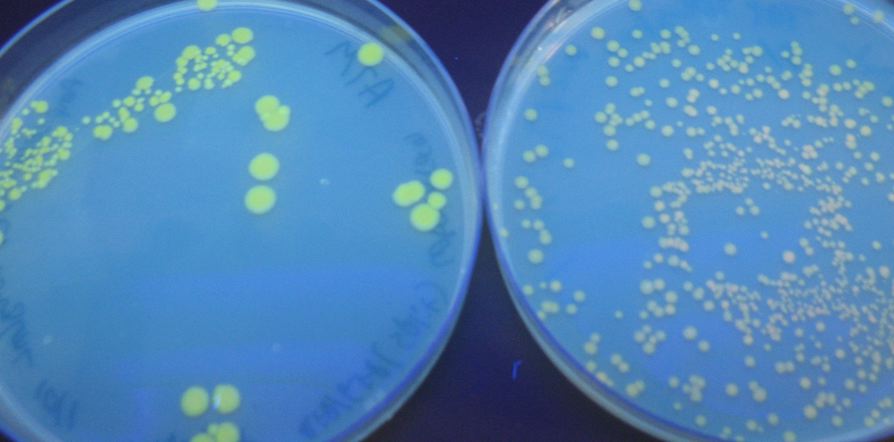
We were able to split two genes: GFP and RFP. Split GFP has strong green fluorescence. Split RFP's red color is much reduced compared to wild-type RFP. It takes overnight incubation at room temperature for the red color to be visible in white light. Both colors are fluorescent under UV light, although the green color predominates. In the first image below, a negative-control (on the left) does not fluoresce, but split GFP (on the right) does.
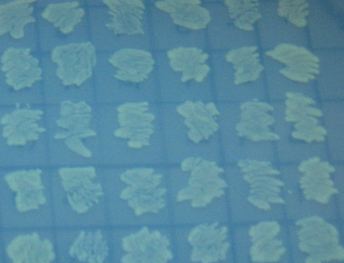
We also wanted to test fluorescence phenotypes with both proteins in the same cells. Below on the left are cells containing both split GFP and split RFP, downstream of the T7 RNA polymerase promoter and co-transformed with T7 RNA polymerase plasmid. These cells look bright green, but the red color is hard to see under UV light. Under white light, the green color is not apparent, but the cells are a little pink. The cells only containing split RFP, on the right, are clearly pink under UV light. However, not all cells are equally colored - some are greener than others, which is a curious result as the cells do not contain GFP of any form.
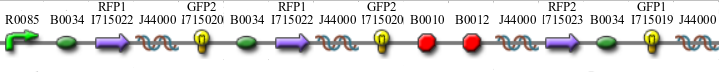
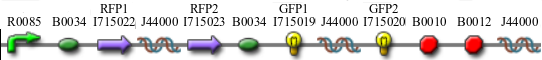
In addition to the regular split GFP and split RFP constructs, we also wanted to test some "hybrid" constructs to ensure that they would not fluoresce. GFP and RFP have similar structures and functions. Previous studies have shown that it is possible to modify GFP to display a wide range of color phenotypes. We created the following parts: Plac-RBS-RFP1-hixC-GFP2 and Plac-RBS-GFP1-hixC-RFP2. A plasmid may, at some point during its flipping process, contain such sequences. We tested to make sure that the similarity of these two proteins did not make them compatible enough to fluoresce. It was found that neither of these parts show any fluorescence or color change.
Constructs
Once we managed to split two genes we proceeded to implement two different graphs in plasmids.
We built the following constructs:
Part Flipping and Computation
Once we had established the creation of successful split reporter genes it became possible to begin constructing HPP computers.