ETHZ
From 2007.igem.org
Stefan Luzi (Talk | contribs) (→Testing new wiki look) |
(Moved new page (former Wiki_rel)) |
||
| Line 1: | Line 1: | ||
| - | [[Image: | + | <center>[[Image:Eth_zh_logo_4.png|900px]]</center> |
| - | + | ||
| - | + | ||
| - | + | <center>[[ETHZ/Main_page | Main Page]] [[ETHZ/Biology | Biology Pespective]] [[ETHZ/Engineering | Engineering Perspective]] [[ETHZ/Meet_the_team | Meet the Team]] [[ETHZ/Internal | Team Notes]] [[ETHZ/Pictures | Pictures!]]</center> | |
| - | < | + | |
| - | + | ||
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | + | __NOTOC__ | |
| - | < | + | <center><font size = '+2'><b> .:: ETH Zurich - EducatETH ''E.coli'' ::. </b></font></center><br> |
| - | + | ||
| - | + | =='''.:: Introduction ::.'''== | |
| - | [ | + | <p>It is the third time the [http://www.ethz.ch ETH Zurich] takes part in [https://2007.igem.org/wiki/index.cgi International Genetically Engineered Machine Competition] (iGEM). This year, our combined team of biologists and engineers has undertaken the task of educating <i>E.coli</i> ! More specifically, in our project (Fig. 1) [[Image:Educateth_Ecoli.png|thumb|The ETH Zurich team undertook the task of engineering a learning mechanism for <i>E.coli</i>. '''(Fig. 1)'''|500px]] we are trying to create <i>E.coli</i> which have the ability to distinguish between two chemicals they may be exposed to after they have undergone a training phase.</p> |
| - | < | + | <p>Stay in this page for an overview of how EducatETH <i>E.coli</i> works, the motivation behind it and its possible future applications. If you want to see the biological design of our system and the parts that it consists of, or if you are interested in building it yourself and want to read the lab protocols, the [[ETHZ/Biology | Biology Perspective]] will be of interest to you. If you want to know more on how EducatETH <i>E.coli</i> has been modeled and simulated, or on its equivalences to systems such as flip-flops and finite state machines, please visit the [[ETHZ/Engineering | Engineering Perspective]]. You may also want to visit [[ETHZ/Meet_the_team | Meet the Team]] for information regarding the team and [[ETHZ/Pictures | Pictures!]] for our photo gallery. Finally, in [[ETHZ/Internal | Team Notes]] you can read the notes exchanged by the team during preparation for the competition.</p> |
| - | + | ||
| - | == | + | ====='''.:: Team Members ::.'''===== |
| - | + | ||
| - | + | ||
| - | + | <p>As [https://2007.igem.org/wiki/index.cgi iGEM] is a synthetic biology competition, the ETH Zurich team consists of balanced numbers of biology and engineering students. Our team members are: </p><ul> | |
| - | + | <li><i>Project advisors</i>: [https://2007.igem.org/User:sven Sven Panke], Joerg Stelling | |
| - | + | <li><i>Undergraduate students</i>: [https://2007.igem.org/User:brutsche Martin Brutsche], [https://2007.igem.org/User:kdikaiou Katerina Dikaiou], [https://2007.igem.org/User:Raphael Raphael Guebeli], [https://2007.igem.org/User:hoehnels Sylke Hoehnel], [https://2007.igem.org/User:LiNan Nan Li], [https://2007.igem.org/User:Stefan Stefan Luzi] | |
| - | + | <li><i>Graduate students</i>: [http://christos.bergeles.net Christos Bergeles], [http://www.tik.ee.ethz.ch/~sop/people/thohm/ Tim Hohm], [http://fm-eth.ethz.ch/eth/peoplefinder/FMPro?-db=phonebook.fp5&-format=pf%5fdetail%5fde.html&-lay=html&-op=cn&Typ=Staff&Suche%5fText=kemmer&Suche%5fText%5fpre=kemmer&-recid=3772770936&-find=/ Christian Kemmer], [https://2007.igem.org/User:JoeKnight Joseph Knight], [https://2007.igem.org/User:uhrm Markus Uhr], [http://www.ricomoeckel.de Rico Möckel] | |
| - | + | </ul> | |
| - | + | <p>For more information on the team members, follow the links or visit [[ETHZ/Meet_the_team | Meet the Team]].</p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | ====='''.:: Motivation ::.'''===== |
| - | === | + | <p> Learning plays a major role for living organisms, as it enables them to survive by adapting to an ever-changing environment. Engineering a simple biological system which exhibits learning behavior is of great interest, as it can support our understanding of this procedure by comparison with natural systems. On the other hand, learning and memorizing plays an equally big role in engineering; from handwriting recognition on PDAs to plain logical circuits storing their binary state on computers, it can be found on numerous everyday life applications. Constructing a biological analogue of a simple memory as known from logic design can hopefully function as a biological building block from which more complex systems may be constructed. We therefore think that EducatETH <i>E.coli</i> operates on an exciting interface between engineering and biology.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ====='''.:: <i>E.coli</i> Intelligence (E.I) - Specifications and analogies ::.'''===== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p>How can we make <i>E.coli</i> bacteria able to report us about their environment? Can we teach bacteria to tell if they have seen before a specific chemical? [[Image:System_blended4.png|thumb|350px| '''Fig. 2: '''EducatETH E. Coli System ]]</p> | |
| - | + | <p>This problem is essentially broken down to constructing a toggle switch which can maintain the state it acquired during a training phase. In Logic Design, this is done using a JK flip-flop with a latch. With this approach, reporting in the testing phase may be implemented with AND gates using the state of the toggle and the current chemical as inputs.<br> | |
| - | + | (Want to read more about this? Visit the [[ETHZ/Engineering| Engineering Perspective]]!)</p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p>Back to biology, a toggle switch has been successfully implemented in [1]. This toggle switch, however, changes states according to one input only as it has one operator site and therefore loses its previous state whenever the system is exposed to a different chemical. This means that it cannot memorize information. To overcome this, we modified the toggle switch in [1] using two operator sites. In this way, a second chemical acting as a “helper” substance present only in the training phase may be used so that the toggle maintains its state during testing.<br> | |
| + | (Want to see the biological design of our modified toggle? Visit the [[ETHZ/Biology | Biology Perspective]]!)</p> | ||
| - | == | + | ====='''.:: A short system description ::.'''===== |
| - | + | <p>EducatETH <i>E.coli</i> is able to recognize between two chemical substances (aTC and IPTG) it has previously been exposed with help of an external chemical signal (AHL). | |
| - | + | In the first part of the training phase (“meeting”), the system is exposed to one of the two chemicals (aTC and IPTG) and AHL is added, causing a steady system behavior. In the second part of the training phase (“memorizing”), all chemicals but AHL are removed, allowing the system to maintain its state. Finally, in the testing phase (“recognizing”), the system is exposed to any of the two chemicals again. Via comparison of the toggle's steady state with the system response the new chemical causes, the system recognizes if it has been exposed to this chemical before or not. As seen on Table , four system states are possible in the testing phase. We chose to use two reporters to control which chemical EducatETH E.coli is exposed to during the training phase (CFP for , YFP for ) and two others to control whether the system sees the same chemical as it did in the training phase (GFP) or not (RFP). Thus, the system response is determined uniquely. </p> | |
| - | |||
| - | + | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | |
| + | ! Training <br>chemical | ||
| + | ! Testing <br>chemical | ||
| + | ! Fluorescence <br>during training | ||
| + | ! Fluorescence <br> during testing | ||
| + | |- | ||
| + | | aTc | ||
| + | | aTc | ||
| + | | CFP or YFP? | ||
| + | | GFP | ||
| + | |- | ||
| + | | aTc | ||
| + | | IPTG | ||
| + | | CFP or YFP? | ||
| + | | RFP | ||
| + | |- | ||
| + | | IPTG | ||
| + | | aTc | ||
| + | | CFP or YFP? | ||
| + | | RFP | ||
| + | |- | ||
| + | | IPTG | ||
| + | | IPTG | ||
| + | | CFP or YFP? | ||
| + | | GFP | ||
| + | |} | ||
| - | + | ====='''.:: References ::.'''===== | |
| + | [1]: Toggle Switch | ||
| - | + | ====='''.:: Acknowledgements ::.'''===== | |
| + | <ul> | ||
| + | <li><p>[http://europa.eu The European Union]</p> | ||
| + | <li><p>[http://www.ethz.ch The ETH Zurich]</p> | ||
| + | <li><p>[http://www.geneart.com GENEART]</p> | ||
| + | </ul><br> | ||
| + | ====='''.:: Links ::.'''===== | ||
| + | <ul> | ||
| + | <li><p>[https://2006.igem.org/wiki/index.php/ETH_Zurich_2005 The ETH Zurich 2005 project]</p> | ||
| + | <li><p>[https://2006.igem.org/wiki/index.php/ETH_Zurich_2006 The ETH Zurich 2006 project]</p></ul><br> | ||
| + | =='''.:: To Do ::.'''== | ||
| + | ====='''.:: New ones ::.'''===== | ||
| - | + | <p><ul> | |
| - | + | <li> Change banner on top of page. | |
| + | <li> Decide on headings type. I like larger headings more, I also like the horizontal line because it separates, put it only on Introduction for you to see how it is. | ||
| + | <li> Possibly put table of contents. | ||
| + | <li> Possibly take EducatETH <i>E.coli</i> from Figure. | ||
| + | <li> Change Figure 2. | ||
| + | <li> Possibly change figure caption format from xxx (Fig. x) to Fig.x: xxx. Put second format on Figure 2 for all to check. | ||
| + | <li> Fix table showing possible system states and reporters. Maybe with some color? | ||
| + | <li> Put cross-reference on table. | ||
| + | <li> Align table left | ||
| + | <li> Add which reporters do what in paragraph. | ||
| + | <li> Put (compressed!) team photo instead of Fig.1 | ||
| + | <li> Fix paragraph with system description, looks somehow bulky. Maybe italics or bold on important things? | ||
| + | <li> Fix reference on toggle switch. | ||
| + | <li> Possibly remove Introduction heading altogether, and just start with the text. | ||
| + | </ul></p> | ||
| - | === | + | ====='''.:: Old ones ::.'''===== |
| - | + | <p><ul> | |
| - | + | <li>Need picture: Bacteria red, Bacteria Green, two pictures showing different exposure to chemicals (''Sylke: see my presentation Sven will hold for me tonight (20.09) -> the FACS guy wants probes of our XFP expressing E.coli - the one who brings it to him can just put it under the microscope and take a few pictures for the wiki'') | |
| - | + | <li>Need picture: Einstein ecoli (''Sylke: does Stefan have the layer file?'') | |
| - | + | <li>Stupidity: All E. Colis are equal, but our E. Colis are more equal than the others :D | |
| - | + | <li>''Katerina'': 2. If we have a separate "Meet the team" page (which is good that we have), we have to make sure that all data about each one of us (short bio+photo) appear also on each one's user page-the same version would be the best, in my opinion. | |
| - | + | <li>''Katerina'': 3. Figure 2, bottom right part needs to be a bit larger/more clear, in my opinion, as it's important. (Christos: If you click, then it becomes larger. Will have the same at the bio part as well... Should I make it bigger anyway? I will change it, it is wrong anyway :D) | |
| - | + | </ul></p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | = | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 14:54, 15 October 2007

.:: Introduction ::.
It is the third time the ETH Zurich takes part in International Genetically Engineered Machine Competition (iGEM). This year, our combined team of biologists and engineers has undertaken the task of educating E.coli ! More specifically, in our project (Fig. 1)
we are trying to create E.coli which have the ability to distinguish between two chemicals they may be exposed to after they have undergone a training phase.Stay in this page for an overview of how EducatETH E.coli works, the motivation behind it and its possible future applications. If you want to see the biological design of our system and the parts that it consists of, or if you are interested in building it yourself and want to read the lab protocols, the Biology Perspective will be of interest to you. If you want to know more on how EducatETH E.coli has been modeled and simulated, or on its equivalences to systems such as flip-flops and finite state machines, please visit the Engineering Perspective. You may also want to visit Meet the Team for information regarding the team and Pictures! for our photo gallery. Finally, in Team Notes you can read the notes exchanged by the team during preparation for the competition.
.:: Team Members ::.
As iGEM is a synthetic biology competition, the ETH Zurich team consists of balanced numbers of biology and engineering students. Our team members are:
- Project advisors: Sven Panke, Joerg Stelling
- Undergraduate students: Martin Brutsche, Katerina Dikaiou, Raphael Guebeli, Sylke Hoehnel, Nan Li, Stefan Luzi
- Graduate students: Christos Bergeles, Tim Hohm, Christian Kemmer, Joseph Knight, Markus Uhr, Rico Möckel
For more information on the team members, follow the links or visit Meet the Team.
.:: Motivation ::.
Learning plays a major role for living organisms, as it enables them to survive by adapting to an ever-changing environment. Engineering a simple biological system which exhibits learning behavior is of great interest, as it can support our understanding of this procedure by comparison with natural systems. On the other hand, learning and memorizing plays an equally big role in engineering; from handwriting recognition on PDAs to plain logical circuits storing their binary state on computers, it can be found on numerous everyday life applications. Constructing a biological analogue of a simple memory as known from logic design can hopefully function as a biological building block from which more complex systems may be constructed. We therefore think that EducatETH E.coli operates on an exciting interface between engineering and biology.
.:: E.coli Intelligence (E.I) - Specifications and analogies ::.
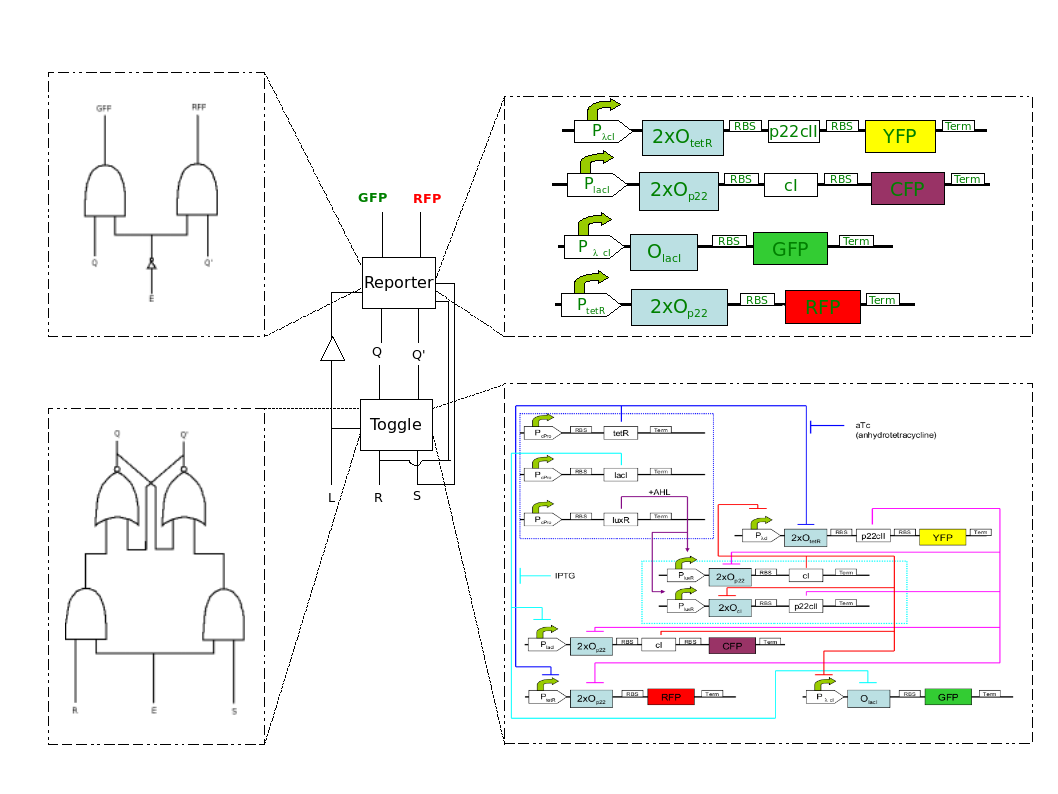
How can we make E.coli bacteria able to report us about their environment? Can we teach bacteria to tell if they have seen before a specific chemical?
This problem is essentially broken down to constructing a toggle switch which can maintain the state it acquired during a training phase. In Logic Design, this is done using a JK flip-flop with a latch. With this approach, reporting in the testing phase may be implemented with AND gates using the state of the toggle and the current chemical as inputs.
(Want to read more about this? Visit the Engineering Perspective!)
Back to biology, a toggle switch has been successfully implemented in [1]. This toggle switch, however, changes states according to one input only as it has one operator site and therefore loses its previous state whenever the system is exposed to a different chemical. This means that it cannot memorize information. To overcome this, we modified the toggle switch in [1] using two operator sites. In this way, a second chemical acting as a “helper” substance present only in the training phase may be used so that the toggle maintains its state during testing.
(Want to see the biological design of our modified toggle? Visit the Biology Perspective!)
.:: A short system description ::.
EducatETH E.coli is able to recognize between two chemical substances (aTC and IPTG) it has previously been exposed with help of an external chemical signal (AHL). In the first part of the training phase (“meeting”), the system is exposed to one of the two chemicals (aTC and IPTG) and AHL is added, causing a steady system behavior. In the second part of the training phase (“memorizing”), all chemicals but AHL are removed, allowing the system to maintain its state. Finally, in the testing phase (“recognizing”), the system is exposed to any of the two chemicals again. Via comparison of the toggle's steady state with the system response the new chemical causes, the system recognizes if it has been exposed to this chemical before or not. As seen on Table , four system states are possible in the testing phase. We chose to use two reporters to control which chemical EducatETH E.coli is exposed to during the training phase (CFP for , YFP for ) and two others to control whether the system sees the same chemical as it did in the training phase (GFP) or not (RFP). Thus, the system response is determined uniquely.
| Training chemical | Testing chemical | Fluorescence during training | Fluorescence during testing |
|---|---|---|---|
| aTc | aTc | CFP or YFP? | GFP |
| aTc | IPTG | CFP or YFP? | RFP |
| IPTG | aTc | CFP or YFP? | RFP |
| IPTG | IPTG | CFP or YFP? | GFP |
.:: References ::.
[1]: Toggle Switch
.:: Acknowledgements ::.
.:: Links ::.
.:: To Do ::.
.:: New ones ::.
- Change banner on top of page.
- Decide on headings type. I like larger headings more, I also like the horizontal line because it separates, put it only on Introduction for you to see how it is.
- Possibly put table of contents.
- Possibly take EducatETH E.coli from Figure.
- Change Figure 2.
- Possibly change figure caption format from xxx (Fig. x) to Fig.x: xxx. Put second format on Figure 2 for all to check.
- Fix table showing possible system states and reporters. Maybe with some color?
- Put cross-reference on table.
- Align table left
- Add which reporters do what in paragraph.
- Put (compressed!) team photo instead of Fig.1
- Fix paragraph with system description, looks somehow bulky. Maybe italics or bold on important things?
- Fix reference on toggle switch.
- Possibly remove Introduction heading altogether, and just start with the text.
.:: Old ones ::.
- Need picture: Bacteria red, Bacteria Green, two pictures showing different exposure to chemicals (Sylke: see my presentation Sven will hold for me tonight (20.09) -> the FACS guy wants probes of our XFP expressing E.coli - the one who brings it to him can just put it under the microscope and take a few pictures for the wiki)
- Need picture: Einstein ecoli (Sylke: does Stefan have the layer file?)
- Stupidity: All E. Colis are equal, but our E. Colis are more equal than the others :D
- Katerina: 2. If we have a separate "Meet the team" page (which is good that we have), we have to make sure that all data about each one of us (short bio+photo) appear also on each one's user page-the same version would be the best, in my opinion.
- Katerina: 3. Figure 2, bottom right part needs to be a bit larger/more clear, in my opinion, as it's important. (Christos: If you click, then it becomes larger. Will have the same at the bio part as well... Should I make it bigger anyway? I will change it, it is wrong anyway :D)