ETHZ/Biology
From 2007.igem.org

.:: Introduction ::.
Our aim is to engineer a biological system which exhibits learning ability and/or adaptative behavior, i.e. a system which can alter its behavior according to external stimuli. We are interested in such a system, since learning and adaptation are playing major roles in living organisms and machine learning has numerous applications in engineering - it is therefore a fantastic interface between engineering and biology. Possible applications are as exciting as biological memories or self-adaptative systems.
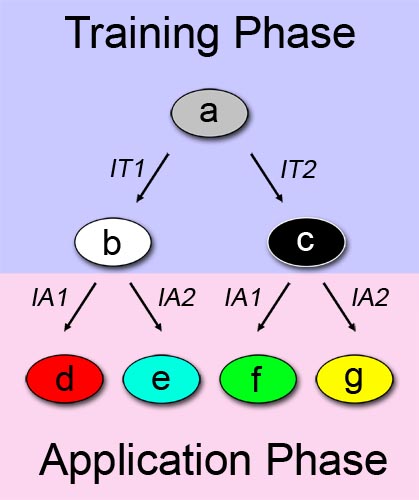
Figure 1 shows a straightforward approach on how to describe learning and adaptive behavior. Imagine there are two phases: a training or learning phase (shown in blue) and an application or "real world" phase (shown in pink). We further assume a system a which can alter its state according to a certain input/stimuli in the first phase. In this binary example, system a changes its state to system b when it is exposed to a first training-phase-input (IT1). Accordingly, system a changes to system c when the other training-phase-input (IT2) is applied.The above described principle is also applicable in the application phase. HOWEVER: Since we have two (b and c), instead of one (a), possible precursor systems, we are ending up in four instead of two different system states. System d is reached, if the first out of two possible application-phase-inputs (IA1) is applied AND system b is the precursor system. This means that in the application phase, two conditions have to be satisfied in order to reach a certain system state. If again AI1 is applied on the precursor system c, the resulting system will be in state f. The same holds true for systems e and g in combination with the second application-phase input (AI2).
How can we now describe learning ability with this approach? The answer is simple: Just define the training-phase-inputs themselves as the information entities to be learned. This also implies that, after the training phase, the information is permanently stored in the system - a memory has been created. According to the intrinsic state of its memory, the system will behave differently when it is exposed to a certain stimuli in a later stage.
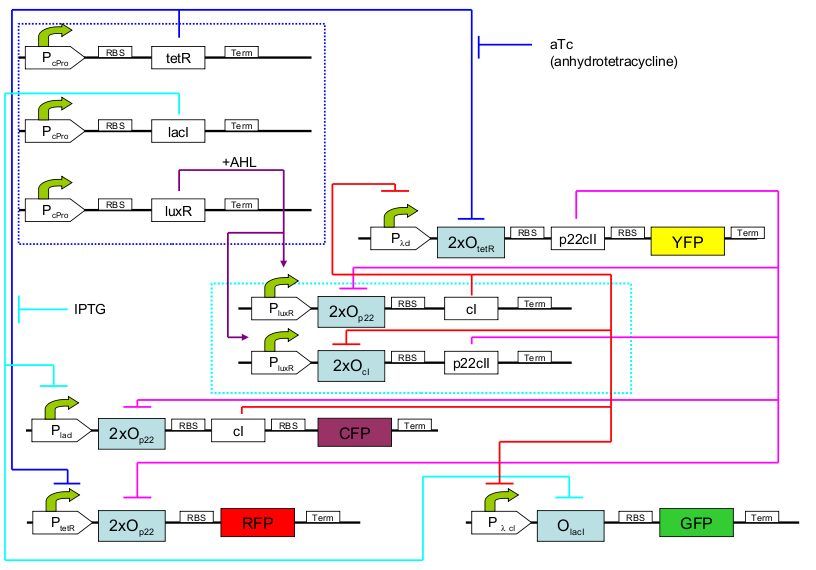
In our project we are constructing an E. coli strain which with the help of an external chemical signal (AHL) is able to remember which of the two chemical substances (aTc and IPTG) it has previously been exposed to. The system architecture is based on a toggle switch consisting of different repressor and activator proteins synthesized from promoters which subject to two different regulations.
In the first operation phase (learning), the system is exposed to one of the two chemicals (aTC and IPTG) and AHL is added, causing a steady system behavior. In the second phase (remembering), the chemicals are removed, but AHL allows the system to still maintain its state.
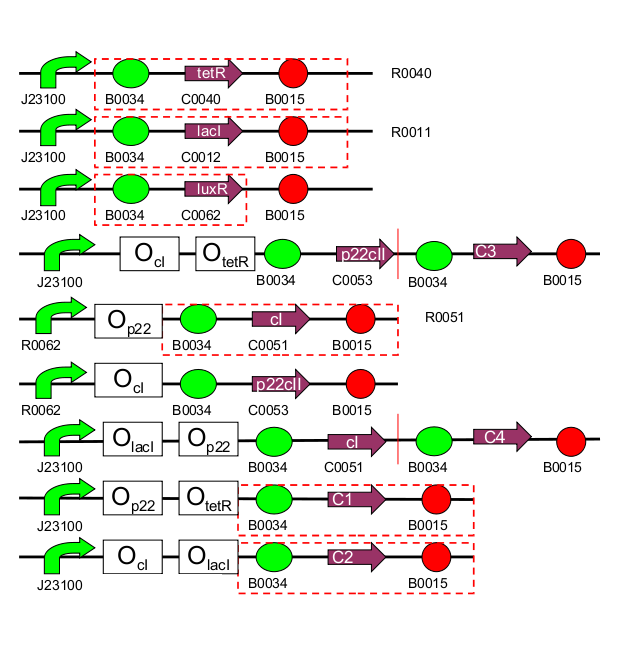
Finally, in the final phase (recognition), the system is exposed to any of the two chemicals again. Its response, reported with 4 fluorescent proteins, differs according not only to which chemical the system is exposed to now, but also to if this chemical is the same that the system has already experienced (learning effect). Therefore, 4 possible system responses are possible: now exposed to aTc and have seen it before/ now exposed to aTc and have not seen it before/ now exposed to IPTG and have seen it before/ now exposed to IPTG and have not seen it before.The systems consists of 11 parts that can be synthesized independently:
| 1 | TetR production (constitutive part of system) |
|---|---|
| 2 | LacI production (constitutive part of system) |
| 3 | LuxR production (constitutive part of system) |
| 4 | 1st half of p22/YFP production (outer part of system, reporting) |
| 5 | 2nd half of p22/YFP production (outer part of system, reporting) |
| 6 | CI production (inner part of system) |
| 7 | p22 production (inner part of system) |
| 8 | 1st half of CI/CFP production (outer part of system, reporting) |
| 9 | 2nd half of CI/CFP production (outer part of system, reporting) |
| 10 | RFP production (reporting) |
| 11 | GFP production (reporting) |
Three plasmids are used to house the above DNA parts, as can be seen from the following table:
| plasmid | resistance | copy type | contents | comments |
|---|---|---|---|---|
| pbr322 | ampicillin | medium | 1,2,3 | constitutive part |
| pck01 | chloramphenicol | low | 4,5,8,9 | outer part |
| pacyc177 | kanamycin | low | 6,7,10,11 | inner part, reporting |
.:: Experiments ::.
.:: References ::.
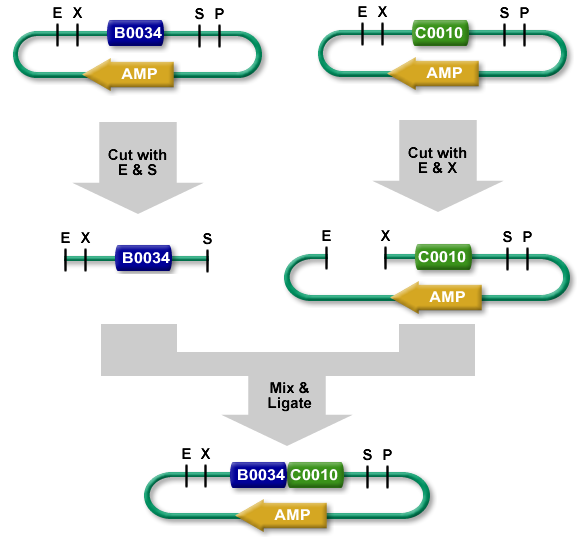
[1] Standard Assembly Process, http://partsregistry.org/Assembly:Standard_assembly
.:: To Do ::.
- Katerina: 1. Number system parts on both figures for easier reference.
- Katerina: 2. Add more info on all system parts and link to the ones existing in the registry. Write info on the ones that didn't exist in the registry (with detailed info such as addition of bp's as Christian and Sven had done).
- Katerina: 3. Add cloning plan. (Christos: Maybe the details will be at the team note's?)