ETHZ/Model
From 2007.igem.org
(text - figure fitting) |
|||
| Line 1: | Line 1: | ||
| - | + | [[Image:Eth_zh_logo_4.png|830px]] | |
| - | + | <!-- | |
<center>[[ETHZ | Main Page]] [[ETHZ/Model | System Modeling]] [[ETHZ/Simulation | Simulations]] [[ETHZ/Biology | System Implementation]] [[ETHZ/Biology/Lab| Lab Notes]] [[ETHZ/Meet_the_team | Meet the Team]] [[ETHZ/Internal | Team Notes]] [[ETHZ/Pictures | Pictures!]]</center><br> | <center>[[ETHZ | Main Page]] [[ETHZ/Model | System Modeling]] [[ETHZ/Simulation | Simulations]] [[ETHZ/Biology | System Implementation]] [[ETHZ/Biology/Lab| Lab Notes]] [[ETHZ/Meet_the_team | Meet the Team]] [[ETHZ/Internal | Team Notes]] [[ETHZ/Pictures | Pictures!]]</center><br> | ||
| + | --> | ||
| + | __NOTOC__ | ||
| + | <html> | ||
| + | <script type="text/javascript" src="http://christos.bergeles.net/eth_dropdowntabs.js"> | ||
| + | |||
| + | /*********************************************** | ||
| + | * Drop Down Tabs Menu- (c) Dynamic Drive DHTML code library (www.dynamicdrive.com) | ||
| + | * This notice MUST stay intact for legal use | ||
| + | * Visit Dynamic Drive at http://www.dynamicdrive.com/ for full source code | ||
| + | ***********************************************/ | ||
| + | |||
| + | </script> | ||
| + | |||
| + | <!-- CSS for Drop Down Tabs Menu #1 --> | ||
| + | <link rel="stylesheet" type="text/css" href="http://christos.bergeles.net/eth_ddcolortabs.css" /> | ||
| + | <div id="colortab" class="ddcolortabs"> | ||
| + | <ul> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ" title="Home" rel="dropmenu_home"><span>Home</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model" title="Modeling" rel="dropmenu_modeling"><span>System Modeling</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation" title="Simulations" rel="dropmenu_simulation"><span>Simulations</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology" title="System Implementation" rel="dropmenu_biology"><span>System Implementation</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team" title="Meet the team" rel="dropmenu_meettheteam"><span>Meet the team</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Pictures" title="Pictures!" rel="dropmenu_pictures"><span>Pictures!</span></a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div class="ddcolortabsline"> </div> | ||
| + | |||
| + | |||
| + | <!--1st drop down menu --> | ||
| + | <div id="dropmenu_home" class="dropmenudiv_a"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Introduction">Introduction Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Team_Members">Team Members Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Acknowledgments">Acknowledgments Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Site_Map">Site map Section</a> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <!--2nd drop down menu --> | ||
| + | <div id="dropmenu_modeling" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Introduction">Introduction Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Model_Overview">Model Overview Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Detailed_Model">Detailed Model Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Final_Model">Final Model Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Modeling_Basics">Modeling Basics Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Mathematical_Model">Mathematical Model Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FSM">FSM View Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FlipFlop">Flip-Flop View Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Parameters">Parameters Page</a> | ||
| + | </div> | ||
| + | |||
| + | <!--3rd drop down menu --> | ||
| + | <div id="dropmenu_simulation" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Introduction">Introduction Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Simulation_of_Test_Cases">Test Cases Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Sensitivity_Analysis">Sensitivity Analysis Section</a> | ||
| + | </div> | ||
| + | |||
| + | <!--4th drop down menu --> | ||
| + | <div id="dropmenu_biology" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#Introduction">Introduction Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#The_Complete_System">The Complete System Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#System_Phases">System Phases Section</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/parts">System Parts Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/Lab">Lab Notes Page</a> | ||
| + | </div> | ||
| + | |||
| + | <!--5th drop down menu --> | ||
| + | <div id="dropmenu_meettheteam" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#The_ETH_Zurich_07_Team">The ETH Zurich 07 Team</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#Team_Description">Team Description</a> | ||
| + | </div> | ||
| + | |||
| + | <script type="text/javascript"> | ||
| + | //SYNTAX: tabdropdown.init("menu_id", [integer OR "auto"]) | ||
| + | tabdropdown.init("colortab", 3) | ||
| + | </script> | ||
| + | </html> | ||
__NOTOC__ | __NOTOC__ | ||
Revision as of 22:07, 23 October 2007
Introduction
As previously discussed in the main page, we are interested in designing a system that is able to adapt to its environment. Our ideas were based on discussions about neural networks, and how we can create a biological system that exhibits the behavior of learning, without having to resort to evolutionary processes.
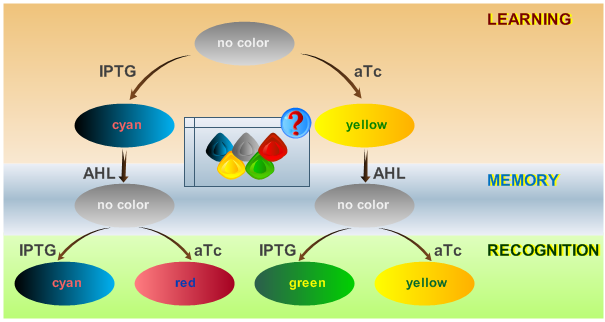
Learning can be considered as a switching of behavior, based on some external stimuli. Thus it comes naturally to work on existing ideas of toggle switches and finite state automatons.
In our system, we are able to distinguish between only two chemicals. We propose only a minimal system that should be able to act as a proof of concept. By introducing the ability to have more final states, in an abstract manner, this correlates with the level of "intelligence" of the biological system. A protocol on how the system should react according to an input is shown in Fig. 1.
The idea behind this protocol is that:
- The system will be able to learn one of two input signals - aTc or IPTG - during a learning phase if no input signal AHL is present. Depending on the input it will report by producing either green or yellow florescence.
- Once the system has learned, the inputs - aTc or IPTG - can be removed and the system goes into a memory state in the presence of the "helper" substance AHL. In this state no output color is reported. Memorizing is guaranteed by removing the input chemicals. This results in a subsequent successful recognition phase.
- During the recognition phase, the inputs aTc or IPTG are (re-)inserted. The system reports by changing its color depending on the input and its current memory state. This is why the system has different florescent properties even in the presence of the same input. The recognition phase takes place in the presence of AHL to keep the memory enabled and avoid another learning phase. Since we would like to separate four different end states for our system, we had to use four fluorescent proteins to encode them.
Model Overview
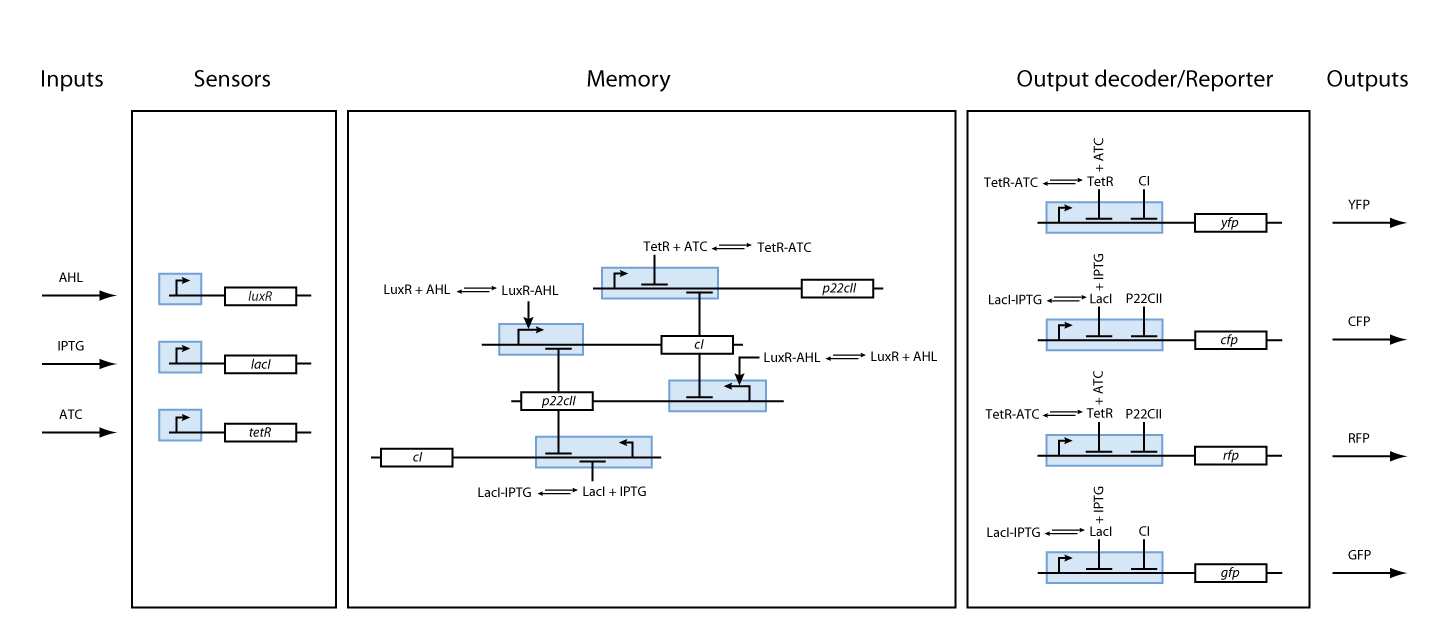
One can start developing our system with a top-down approach. We start with the classical black box approach as shown in Figure 2.
Basedcan be summarized as followshas can be summarized as follows:
- Two inputs to be learned/detected/adapted to,
- One input to switch on the memory,
- To alternate between at least three states. Hence why we decided to use two state variables - cI and p22cII.
- Four florescent signals for the outputs. One could also decide to take six output signals into account, to further distinguish the learning phase from the recognition phase. However, we restricted ourselves to 4 outputs to reduce the number of genes that are needed to implement the signals.
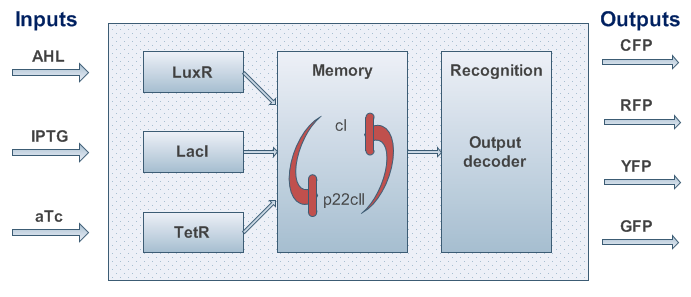
Based on the above, the internal structure of the system can be defined, and seen in Figure 3. However, we had to keep in mind that the proposed system should be implemented in DNA, and that it would be sensitive to noise. As a result, we took several actions to achieve better experimental results and easier DNA construction:
- To be more robust against perturbations, we coupled the state variables cI and p22cII in the way that is well known from memory circuits. Based on this approach, one state variable is depressing the other one, and the system's internal toggle has the possibility of reaching two stable states.
- Since - due to their size - proteins can only hardly pass the cell membrane (if they are not actively transported through the cell membrane), we decided to use the much smaller inducer molecules AHL, IPTG and aTc as inputs. However, since these inducers cannot directly act on the transcription of the DNA nor on the production of proteins, we need to produce the sensor proteins LuxR, LacI and TetR that build complexes with AHL, IPTG and aTc, respectively.
- The sensor proteins and complexes are used to control the memory formation and the production of the florescent reporter proteins CFP, RFP, GFP and YFP.
Detailed Model
In order to test our ideas, we had to find the equations that describe the proposed model, and fill in the gaps. In this section, we provide the details of our model, giving precise descriptions of the involved molecules and proteins. Our model is created on the basic principles of finite state machines. It is a biological automaton that moves from the learning states, to the memorizing and recognizing states, as it was presented in Fig. 1. For a detailed analysis of the underlying finite state machine please see the section Finite State Machine View of the System. To see how our memory system translates into a flip flop memory have a look at the section An Engineer's View on Biological Learning
Sensors
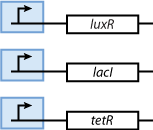
As we can observe in Fig. 3, our system is composed from three basic subparts. The first part contains the sensors. Our sensors are the proteins LacI, luxR and TetR, and they are constitutively produce, in order to regulate the operation of the rest of the system. The sensing subsystem can be seen in Fig. 4.
Memory
The second subsystem is responsible for the creation and control of memories. The memory control is based on the following underlying mechanisms:
- The sensor proteins form complexes together with the inducers. These complexes are used to either activate (in case of the complex consisting of luxR and AHL) or repress (in case of the complexes consisting of LacI and IPTG as well as TetR and aTc) the DNA transcription of the proteins cI and p22cII.
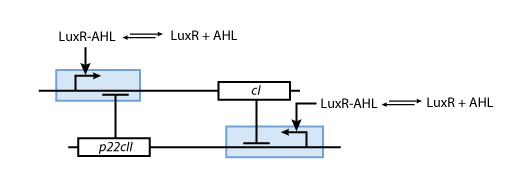
- p22cII and cI repress the DNA transcription of each other, so that the closed loop system behaves as a toggle; a dynamic system with only two possible steady states (see Fig. 6).
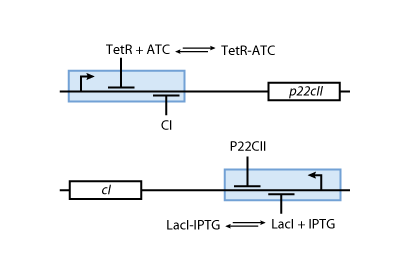
- Fig. 5 shows the protein production system that is used during the learning phase. During the learning phase, there is still no cI or p22cII produced. They are produced, only if either IPTG or aTc is added, respectively. Since no AHL is present, the inner toggle switch (see Figure 6) is turned off.
- During the memory phase, AHL is added and the IPTG and aTc are removed. That is why only the inner toggle switch (see Fig. 6) is turned on while the protein production systems shown in Fig. 5 are deactivated. Depending on what was produced during the learning phase, the production of either cI or p22cII is continued. That is why the system can act as memory, effectively storing the information that it is exposed to.
Based on all the above, we present the final assembly of the memory subsystem in Fig. 7.
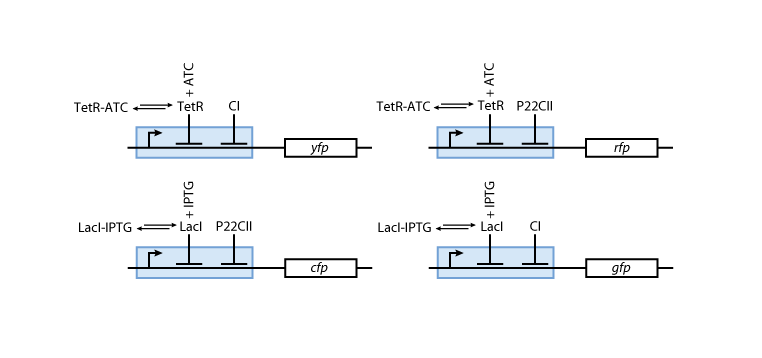
Reporters
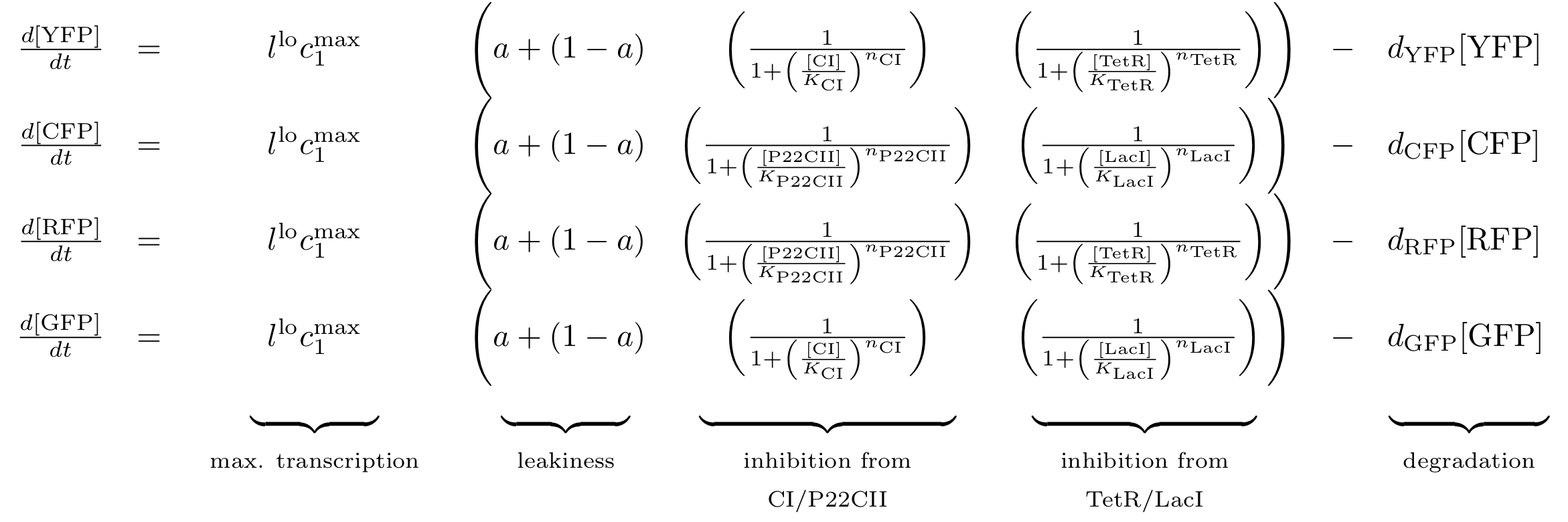
Fig. 8 gives an overview about the reporter subsystem. Florescent reporter proteins are expressed depending on the inducer concentrations, and the concentrations of cI and p22cII. For example, the presence of either TetR or cI will repress the production of YFP. However, if the inducer aTc is present, aTc will bind to TetR which can no longer block the production of YFP. We are using four fluorescent proteins, to encode the steady states of our system at the final recognition stage. In this way, we are able to distinguish between all the different transition paths of our biological automaton.
Final Model
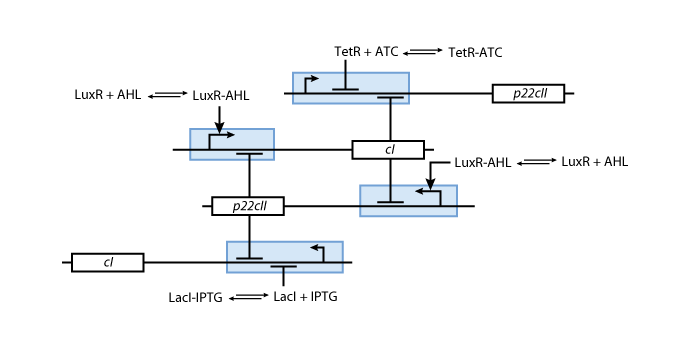
We have so far presented all the parts that are needed in order to model and simulate the behavior of a biological automaton with the ability to memorize and recognize the chemical that it is exposed to. By following the details presented in the previous section, we have all the necessary information to fully understand the interior of the black boxes that were presented in Fig. 2 and Fig. 3. Our overall system model is presented in Fig. 9.
Mathematical Model
Based on the modeling done so far, we can derive the equations that govern the behavior of our system. The model is governed by sets of coupled ordinary differential equations which are presented below. We use a simple notation for the different elements of the equations. Namely:
- All concentrations are given in brackets (for example [cI]).
- All decay constants are described by a variable d followed by the name of the protein they refer to.
- The production of the proteins is described by a basic constant production level that models the leak of the production system, and a factor of l and cmax that describe the maximum production of a protein, given in [M].
- Depending on whether the DNA for a protein is implemented on a low or a high copy plasmid, we distinguish between llo and lhi, respectively.
For a more basic introduction into how we transferred our model into equations, see the section Modeling Basics.
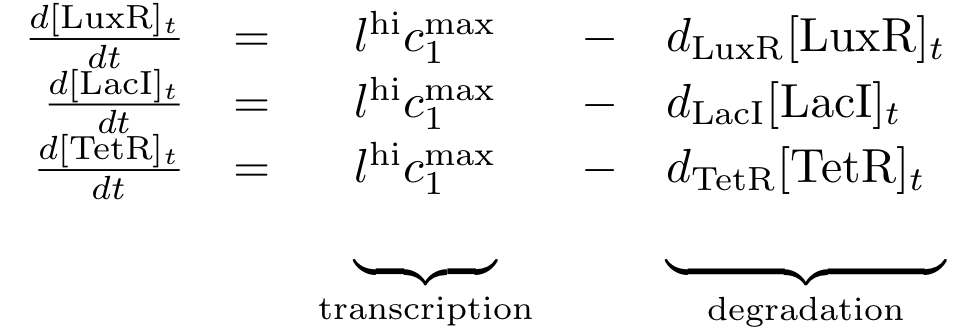
Constitutively produced proteins
The equations for the constitutively produced proteins are very simple, since there is no dependence on other proteins. They are designed so that the protein concentration reaches the value lhi*cmax/d at steady state.
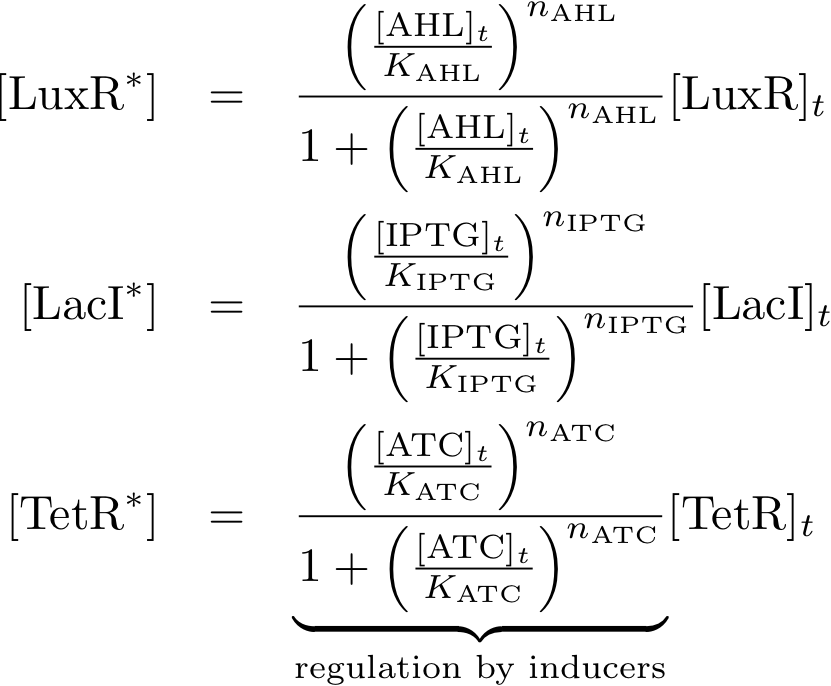
Allosteric regulation
These equations describe the formation of complexes between the inducers and sensor proteins. We do not use differential equations, but describe directly the concentrations of the complexes. This is a valid assumption, provided that we always wait a sufficient time, and the system reaches a steady state. We describe the total amount of proteins with the index 't', while we use the index '*' for proteins that build a complex with their respective inducer. For example:
- [TetRt] describes the total amount of TetR that is available.
- [TetR*] describes the proteins that are available as a complex with aTc, and
- [TetR] gives the concentration of free TetR proteins.
Learning and memory subsystem
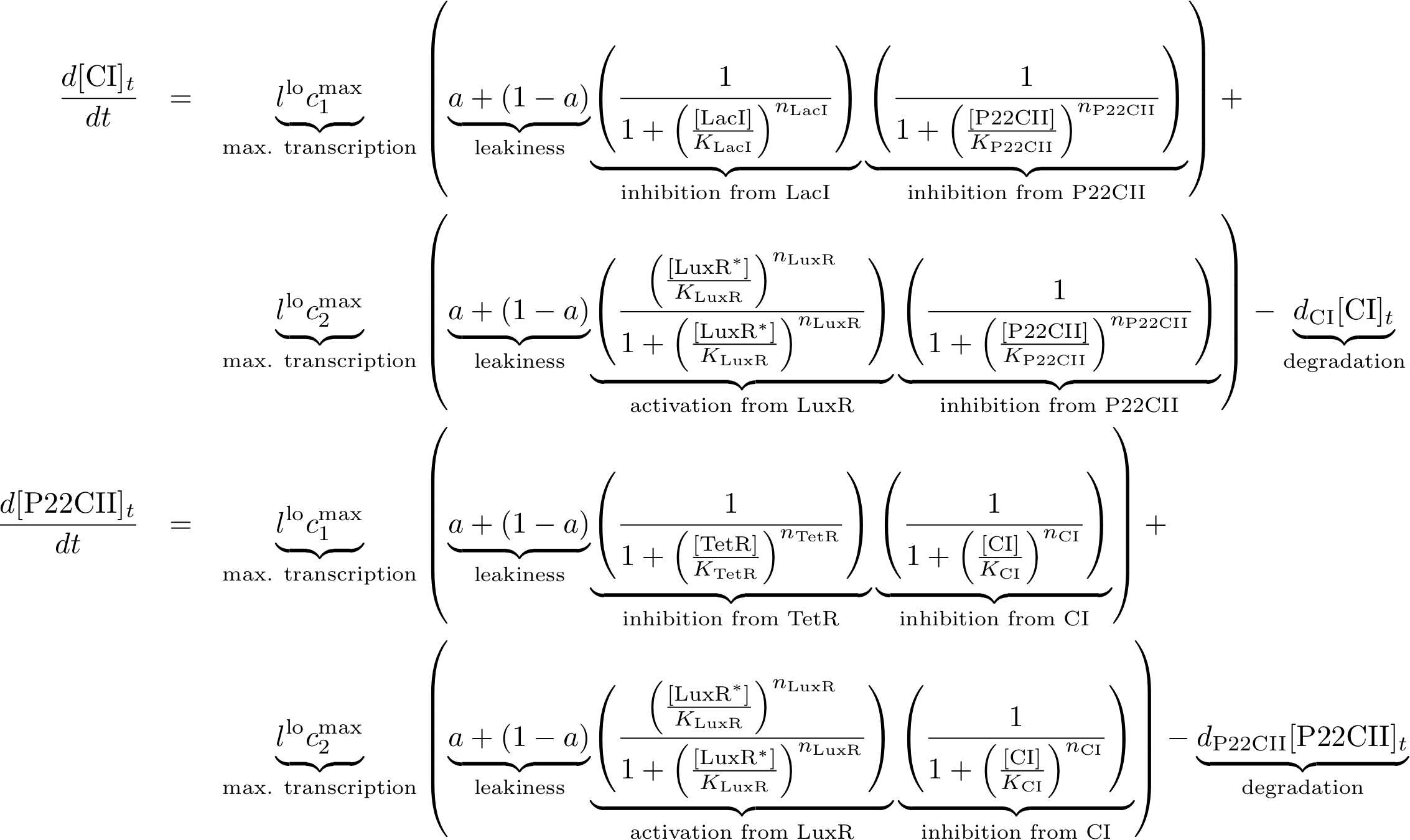
The learning and memory subsystem is the core of the system that we are trying to model and implement. It is characterized by the feedback between its state variables/proteins cI and p22cII. Its behavior is further complicated by the variation of the production of the aforementioned proteins because of the inputs. The following equations describe the concentrations of the memory proteins as a system of coupled differential equations. The equations consist of two major production parts and a decay part.
- The first production part models the production of either cI or p22cII during the learning phase, and corresponds to the model in Fig. 5.
- The second production part describes the inner toggle switch that was shown in Fig. 6.
Reporting subsystem
The equations for the reporting subsystem finally describe the production of the florescence proteins depending on the inputs and memory proteins as modeled in Figure 8. Note that both inputs and memory proteins act repressively on the production of the florescence proteins. So e.g. YFP is only produced when there is both no cI and all TetR is bound in a complex together with aTc.
The systems of equations presented above describe and predict the behavior of our system. We have simulated the behavior of our system at steady states, and the results can be seen in the section Simulations. In order to increase the accuracy of our results, we conducted an extensive literature survey, in order to isolate and find the parameters of our system. Since this is a burden for every team undertaking a complicated project in synthetic biology, we are presenting our full table of parameters in the Parameters page.