ETHZ/Modeling Basics
From 2007.igem.org
(→Regulated Protein Production) |
(→Regulated Protein Production) |
||
| Line 32: | Line 32: | ||
=== Inhibition === | === Inhibition === | ||
| - | To derive the equations describing the regulated transcription, we first need a model how the transcription factor interacts with DNA. The most simple model is when the protein binds reversibly to the DNA | + | To derive the equations describing the regulated transcription, we first need a model how the transcription factor interacts with DNA. The most simple model is when the protein binds reversibly to the DNA: |
[[Image:basic_fig05.png]] | [[Image:basic_fig05.png]] | ||
Revision as of 15:22, 7 October 2007
Contents |
Introduction
The functioning of our model depends mostly on protein concentrations. These proteins are produced within the E. coli cells based on genes that we introduced into the cell. To understand the system, it is crucial to model the gene expression and regulation accurately.
This Wiki page is intended to present the basic mechanisms and assumptions that went into the mathematical description of our iGEM model.
Constitutive Protein Production
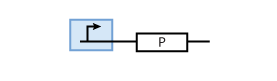
The most simple parts in the system are genes that are continuously transcribed to produce a protein. At the same time, proteins have a certain half-life time, which means that they are degraded. This leads to the following simple model of protein production/degradation shown in Fig. 1.
To find the concentration of protein P (as a function of time), the system of Fig. 1 can be written as an ODE
This equation states that the change of protein concentration is a function of protein production (cmax) and protein degradation (dP[P]).
It is worth looking at the protein production a bit closer. The production of protein P depends on the expression of a gene that codes for this protein. In the case of constant protein production, the gene can be modeled by a constitutive promoter and a coding region for the protein (Fig. 2).
Regulated Protein Production
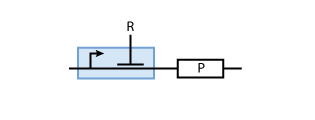
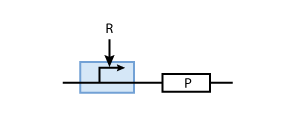
To build a system with logic functionality, it is necessary to produce a protein depending on the concentration of another protein (i.e. a transcription factor). To model such a system, the promoter of the constitutive production system in Fig. 2 must be extended to take into account the presence of the regulatory protein R. There are two possible cases: the regulatory protein either inhibits or activates the expression of the gene (see Figs. 3 and 4, respectively).
Inhibition
To derive the equations describing the regulated transcription, we first need a model how the transcription factor interacts with DNA. The most simple model is when the protein binds reversibly to the DNA:
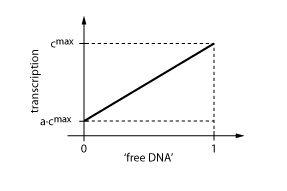
When the transcription factor binds to DNA, it blocks the enzymes transcribing the gene. Thus, the higher the concentration of R, the smaller the transcription of the gene.