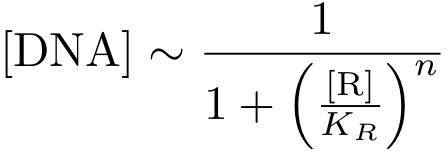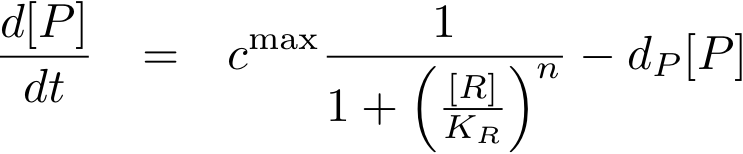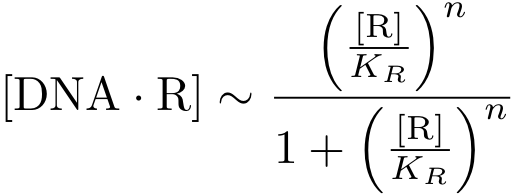ETHZ/Modeling Basics
From 2007.igem.org
(→Inhibition) |
(→Inhibition) |
||
| Line 49: | Line 49: | ||
Now, we have basically everything together to write down a differential equation for the concentration of protein P whose expression is regulated by protein R: | Now, we have basically everything together to write down a differential equation for the concentration of protein P whose expression is regulated by protein R: | ||
| - | [[Image:basic_eq05.png|center| | + | [[Image:basic_eq05.png|center|253px]] |
The transcription does not always take place at the maximum rate ''c''<sup>max</sup>, but is modulated by the concentration of R. | The transcription does not always take place at the maximum rate ''c''<sup>max</sup>, but is modulated by the concentration of R. | ||
| Line 55: | Line 55: | ||
This is not the full story yet, however. Even though inhibition is maximal, in reality, there is still some transcription observable (usually around 10-20% of the maximum transcription rate). This is illustrated in Fig. X. Thus we have to introduce some 'basic transcription' that always takes place: | This is not the full story yet, however. Even though inhibition is maximal, in reality, there is still some transcription observable (usually around 10-20% of the maximum transcription rate). This is illustrated in Fig. X. Thus we have to introduce some 'basic transcription' that always takes place: | ||
| - | [[Image: | + | [[Image:basic_eq06.png|center|300px]] |
Note that the basic transcription rate ''a'' is introduced as a percentage of the maximum transcription rate ''c''<sup>max</sup>. Regulation by protein R now is only effective in the range between ''a''·''c''<sup>max</sup> and ''c''<sup>max</sup>. | Note that the basic transcription rate ''a'' is introduced as a percentage of the maximum transcription rate ''c''<sup>max</sup>. Regulation by protein R now is only effective in the range between ''a''·''c''<sup>max</sup> and ''c''<sup>max</sup>. | ||
=== Activation === | === Activation === | ||
Revision as of 16:37, 7 October 2007
Contents |
Introduction
The functioning of our model depends mostly on protein concentrations. These proteins are produced within the E. coli cells based on genes that we introduced into the cell. To understand the system, it is crucial to model the gene expression and regulation accurately.
This Wiki page is intended to present the basic mechanisms and assumptions that went into the mathematical description of our iGEM model.
Constitutive Protein Production
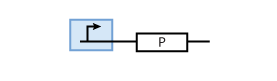
The most simple parts in the system are genes that are continuously transcribed to produce a protein. At the same time, proteins have a certain half-life time, which means that they are degraded. This leads to the following simple model of protein production/degradation shown in Fig. 1.
To find the concentration of protein P (as a function of time), the system of Fig. 1 can be written as an ODE
This equation states that the change of protein concentration is a function of protein production (cmax) and protein degradation (dP[P]).
It is worth looking at the protein production a bit closer. The production of protein P depends on the expression of a gene that codes for this protein. In the case of constant protein production, the gene can be modeled by a constitutive promoter and a coding region for the protein (Fig. 2).
Regulated Protein Production
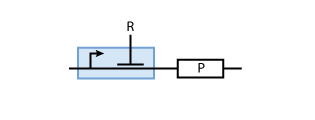
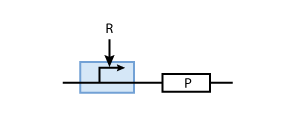
To build a system with logic functionality, it is necessary to produce a protein depending on the concentration of another protein (i.e. a transcription factor). To model such a system, the promoter of the constitutive production system in Fig. 2 must be extended to take into account the presence of the regulatory protein R. There are two possible cases: the regulatory protein either inhibits or activates the expression of the gene (see Figs. 3 and 4, respectively).
Inhibition
To derive the equations describing the regulated transcription, we first need a model how the transcription factor interacts with DNA. The most simple model is when the protein binds reversibly to the DNA:
Note that the equation above involves n transcription factors. For certain transcription factors the number of proteins involved is indeed greater than 1. These are interesting cases that enable applications such as toggle switches.
When the transcription factor binds to DNA, it blocks the enzymes transcribing the gene. Thus, the higher the concentration of R, the smaller the transcription of the gene. By controlling the concentration of the regulatory protein R, the expression of protein P can effectively be regulated.
To understand this process in more detail, we make the simplification that the binding of R to DNA is in equilibrium. That is, one can write
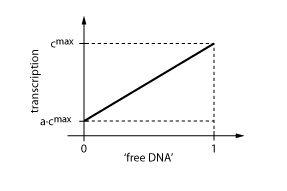
In case of inhibition, the expression of the gene is proportional to the probability that the DNA is 'free' (meaning that there is no transcription factor bound to it). After some algebraic manipulation of the above equation an expression for the 'free DNA' as a function of transcription factor concentration can be derived:
Now, we have basically everything together to write down a differential equation for the concentration of protein P whose expression is regulated by protein R:
The transcription does not always take place at the maximum rate cmax, but is modulated by the concentration of R.
This is not the full story yet, however. Even though inhibition is maximal, in reality, there is still some transcription observable (usually around 10-20% of the maximum transcription rate). This is illustrated in Fig. X. Thus we have to introduce some 'basic transcription' that always takes place:
Note that the basic transcription rate a is introduced as a percentage of the maximum transcription rate cmax. Regulation by protein R now is only effective in the range between a·cmax and cmax.