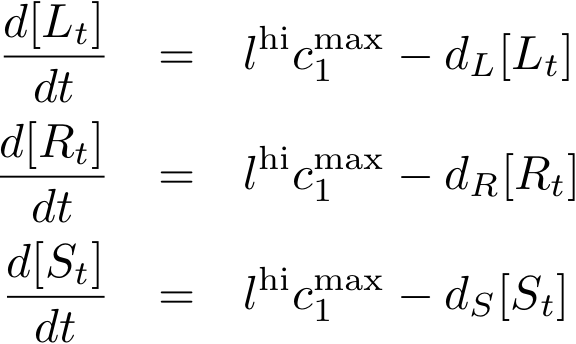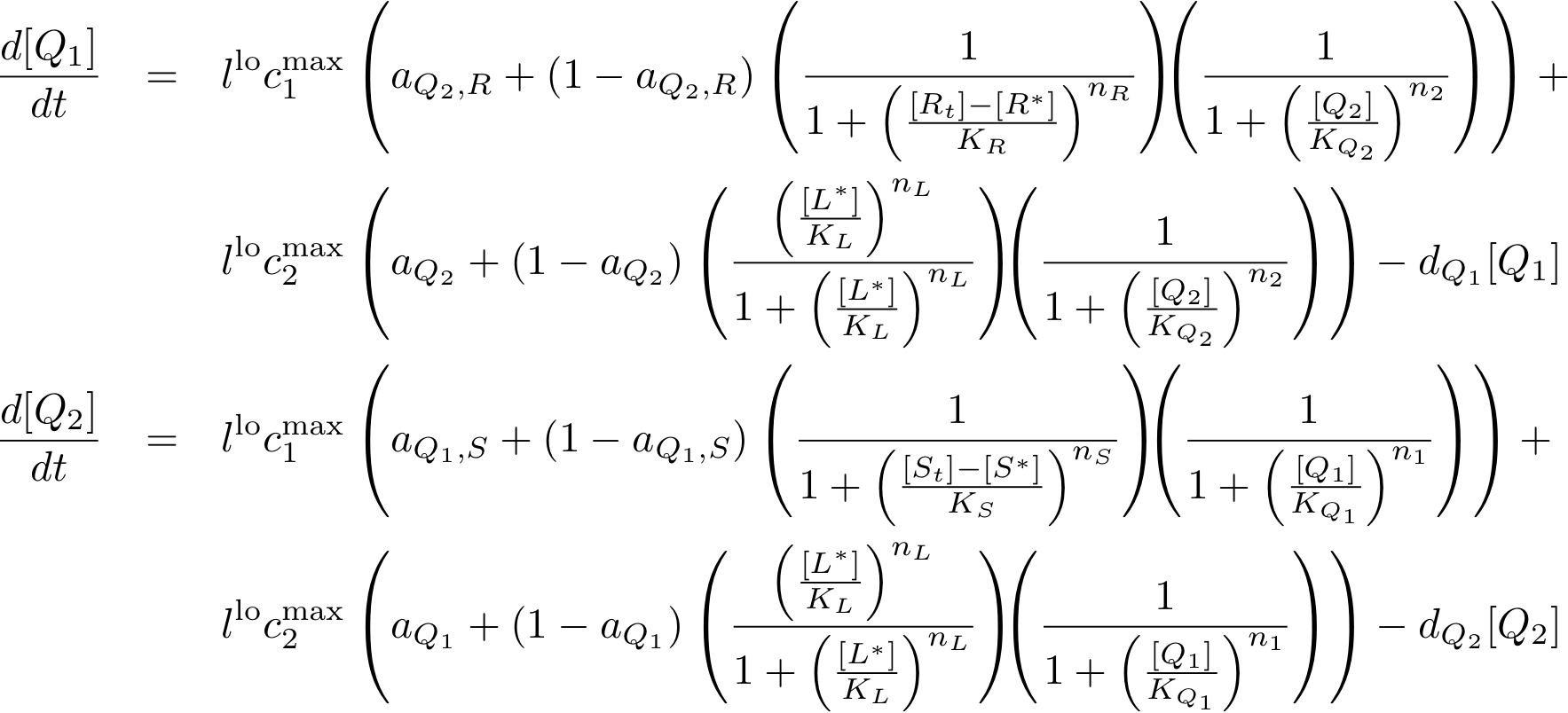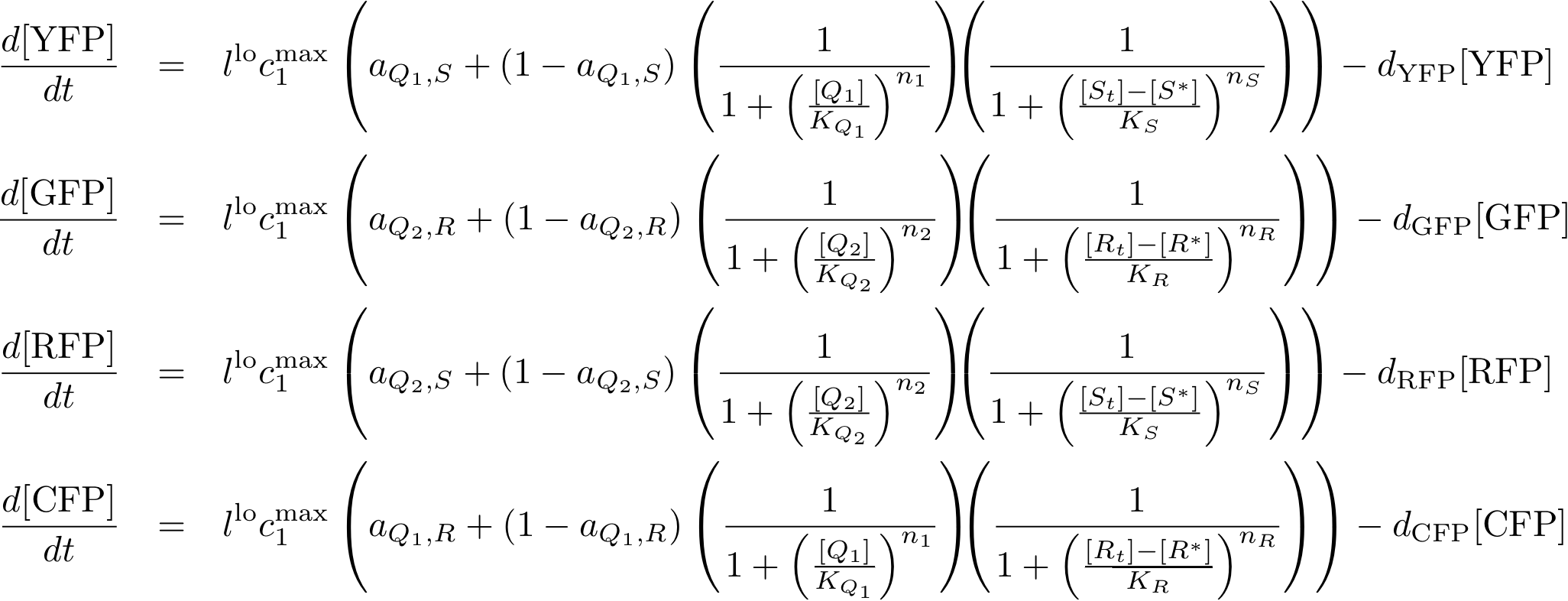ETHZ/Simulations
From 2007.igem.org
(Difference between revisions)
(→Model Parameters) |
(→References) |
||
| Line 194: | Line 194: | ||
# Detailed map of a cis-regulatory input function (http://www.pnas.org/cgi/content/full/100/13/7702?ck=nck) | # Detailed map of a cis-regulatory input function (http://www.pnas.org/cgi/content/full/100/13/7702?ck=nck) | ||
# Parameter Estimation for two synthetic gene networks (http://ieeexplore.ieee.org/iel5/9711/30654/01416417.pdf) | # Parameter Estimation for two synthetic gene networks (http://ieeexplore.ieee.org/iel5/9711/30654/01416417.pdf) | ||
| + | # Supplementary on-line information for "A Synthetic gene-metabolic oscillator" (no link) | ||
== Variable Mapping == | == Variable Mapping == | ||
Revision as of 20:32, 11 September 2007
Contents |
Basic Model
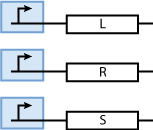
Constitutively produced proteins
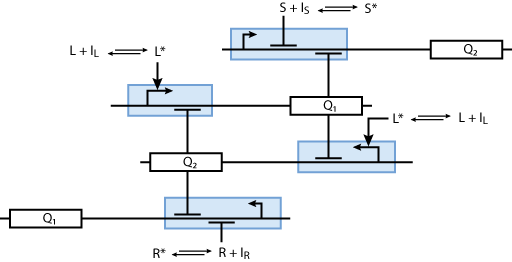
Learning system
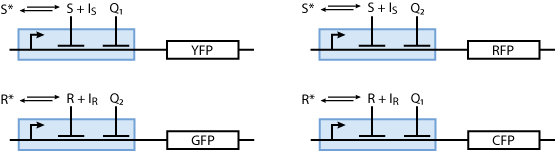
Reporter system
System Equations
Constitutively produced proteins
Learning system
Reporter system
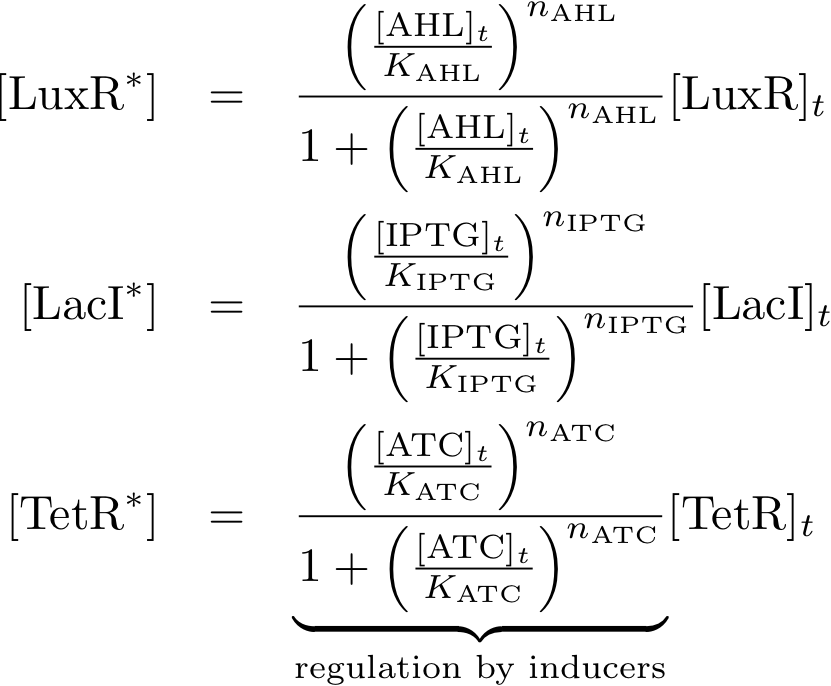
Allosteric regulation
Model Parameters
| Parameter | Value | Description | Comments |
|---|---|---|---|
| aR | basic production of lacI | ||
| aS | basic production of tetR | ||
| aL | basic production of luxR | ||
| aQ1 | basic production of cI | ||
| aQ2 | basic production of p22cII | ||
| aGFP | basic production of GFP | ||
| aRFP | basic production of RFP | ||
| dR | 0.06 | degradation of lacI | Ref. [4] |
| dS | degradation of tetR | ||
| dL | degradation of luxR | ||
| dQ1 | degradation of cI | ||
| dQ2 | degradation of p22cII | ||
| dGFP | degradation of GFP | ||
| dRFP | degradation of RFP | ||
| kR | 0.22 [mM/h] | promoter strength (Promoter R0010) | |
| kS | promoter strength (Promoter R0040) | ||
| kL | promoter strength (Promoter R0062) | ||
| kQ1 | 0.20 [mM/h] | promoter strength (Promoter R0051) | |
| kQ2 | promoter strength (Promoter R0053) | ||
| KR | 1.3e-3 - 2e-3 [mM/h] | lacI repressor dissociation constant | lower value is from Ref. [2], higher value is from Ref. [?] |
| KIR | 1.5e-10 [mM/h] | IPTG-lacI repressor dissociation constant | |
| KS | tetR repressor dissociation constant | ||
| KIS | aTc-tetR repressor dissociation constant | ||
| KL | luxR activator dissociation constant | ||
| KIL | AHL-luxR activator dissociation constant | ||
| KQ1 | 2e-3 [mM/h] | cI repressor dissociation constant | |
| KQ2 | p22cII repressor dissociation constant | ||
| nR | 1 | lacI repressor Hill cooperativity | |
| nIR | 2 | IPTG-lacI repressor Hill cooperativity | |
| nS | 3 | tetR repressor Hill cooperativity | Ref. [3] |
| nIS | aTc-tetR repressor Hill cooperativity | ||
| nL | 1 | luxR activator Hill cooperativity | Ref. [3] |
| nIL | 1 | AHL-luxR activator Hill cooperativity | Ref. [3] |
| nQ1 | 1.9 | cI repressor Hill cooperativity | |
| nQ2 | p22cII repressor Hill cooperativity |
References
- A synthetic time-delay circuit in mammalian cells and mice (http://www.pnas.org/cgi/content/abstract/104/8/2643)
- Detailed map of a cis-regulatory input function (http://www.pnas.org/cgi/content/full/100/13/7702?ck=nck)
- Parameter Estimation for two synthetic gene networks (http://ieeexplore.ieee.org/iel5/9711/30654/01416417.pdf)
- Supplementary on-line information for "A Synthetic gene-metabolic oscillator" (no link)
Variable Mapping
| Variable | Compound |
|---|---|
| R | lacI |
| IR | IPTG |
| S | tetR |
| IS | aTc |
| L | luxR |
| IL | AHL |
| Q1 | cI |
| Q2 | p22cII |