ETHZ/Wiki rel
From 2007.igem.org

====.:: Introduction ::.====
The Federal Institute of Technology (ETH) Zurich is participating at the International Geneticaly Engineered Machines (iGEM) Competition, for the 3rd year . Our combined team of Biologists and Engineers has undertaken the task of "Educating E. Coli" (see Fig. 1).
After elaborating ( :) ) on the definition of learning, we drew ideas from the field of logic design, and we are trying to create E. Coli with the ability to recognize the chemicals that they are exposed to, based on a previous training phase.This page presents the basic ideas of our project. In the Biology Perspective, you can find details on the biological system that we are trying to implement, the parts it consists of, and our biological experiments. In the Engineering Perspective, you can find the governing equations of our biological system, its modeling, and the simulations that we have carried out. In Meet the Team, you can find more information concerning the member of the ETH Zurich team. In Team Notes, you can find notes taken during the preparation process, to facilitate the communication between the team members. Lastly, in Pictures!, you can find our ETH Zurich preparation gallery :).
.:: Team Members ::.
Since this is a Synthetic Biology competition, the ETH Zurich team consists of balanced amounts of Biology students, and Engineering students.
- Project advisors: Sven Panke, Joerg Stelling
- Undergraduate students: Martin Brutsche, Katerina Dikaiou, Raphael Guebeli, Sylke Hoehnel, [http://leemnan.spaces.live.com Nan Li], Stefan Luzi
- Graduate students: [http://christos.bergeles.net Christos Bergeles], [http://www.tik.ee.ethz.ch/~sop/people/thohm/ Tim Hohm], [http://fm-eth.ethz.ch/eth/peoplefinder/FMPro?-db=phonebook.fp5&-format=pf%5fdetail%5fde.html&-lay=html&-op=cn&Typ=Staff&Suche%5fText=kemmer&Suche%5fText%5fpre=kemmer&-recid=3772770936&-find=/ Christian Kemmer], Joseph Knight, Markus Uhr, [http://www.ricomoeckel.de Rico Möckel]
For more information on the team members, follow the links, or visit the Meet the Team web page.
.:: E. Coli Intelligence (E.I) ::.
How can we make E. Coli bacteria able to report us about their environment? Can we teach bacteria to recognize a specific chemical?
Based on these questions we set forth to designing biological circuits to educate E. Coli cells, and to improve the E. Coli Intelligence...Our goal is to design an E.coli strain, able to distinguish between two chemical substances after an assigned learning process. The learning process is induced by an external chemical signal (teaching substance). Afterwards, the bacterial strain will be taken to a testing phase, where the output will be in either green or red fluorescence, depending on whether the bacteria recognized the same chemical substance in the testing phase as it was trained in the learning phase or a different one.
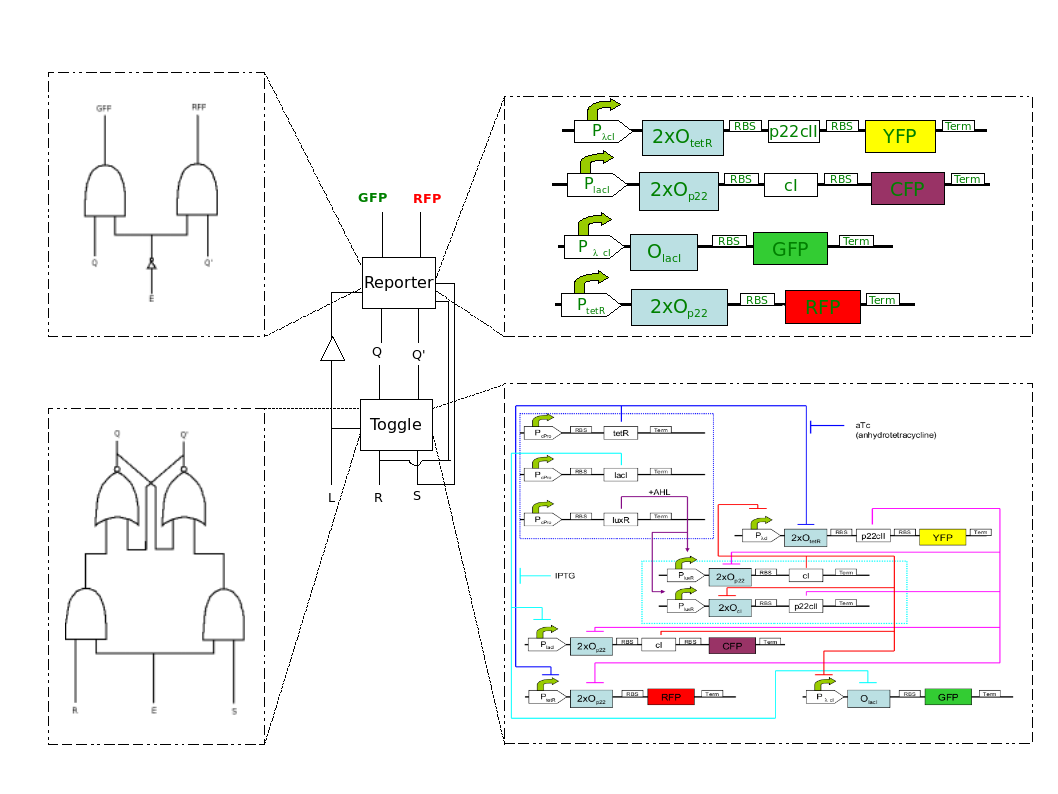
In order to come to this idea for implementation, we had a discussion based on ideas from Logic Design. A toggle that keeps it state, can be a JK flip-flop with a latch. Reporting circuits can be constructing using AND gates, where the inputs are the chemicals and the previous state of the JK flip-flop. In Fig.2, we present the blending of the engineering and biological perspective.
The toggle that our system contains is based on the toggle described in [1]. However, since we want two separate processes, we had to change appropriately the promoter sites. Hence, now, the presence of two chemicals is required for the toggle to function. This allows us to distinguish between the states of learning and recognition. At the first state, due to the learning protein HSL, the toggle is active, and reaches a steady state based on the different input chemicals. At the second state, the reporters are activated, and fluorescent protein is produced based on the newly added chemical, and the former steady state of the toggle. The newly added chemical doesn't affect the toggle, since the learning protein HSL is abscent. In the final system, we have added two more reporters, in order to check which chemical the system is currently learning. These promoters are active both during the learning, and the recognition process.
.:: References ::.
.:: To Do ::.
- Need picture: Bacteria red, Bacteria Green, two pictures showing different exposure to chemicals (Sylke: see my presentation Sven will hold for me tonight (20.09) -> the FACS guy wants probes of our XFP expressing E.coli - the one who brings it to him can just put it under the microscope and take a few pictures for the wiki)
- Need picture: Einstein ecoli (Sylke: does Stefan have the layer file?)
- Stupidity: All E. Colis are equal, but our E. Colis are more equal than the others :D
- Katerina: 2. If we have a separate "Meet the team" page (which is good that we have), we have to make sure that all data about each one of us (short bio+photo) appear also on each one's user page-the same version would be the best, in my opinion.
- Underlined supervisors and stuff: Switched it to italics. Larger heading look bad, I've checked it...
- Katerina: 3. Figure 2, bottom right part needs to be a bit larger/more clear, in my opinion, as it's important. (Christos: If you click, then it becomes larger. Will have the same at the bio part as well... Should I make it bigger anyway? I will change it, it is wrong anyway :D)