ICAM-based Targeting and Signaling
From 2007.igem.org
Amendelsohn (Talk | contribs) |
Amendelsohn (Talk | contribs) |
||
| (One intermediate revision not shown) | |||
| Line 1: | Line 1: | ||
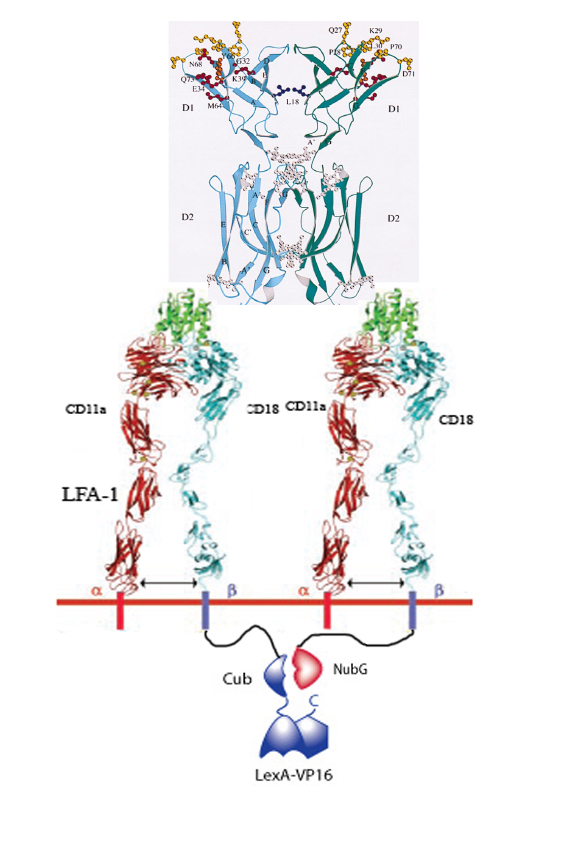
| + | == How it works == | ||
| + | 1. Infarced myocardium induces expression of ICAM. | ||
| + | |||
| + | 2. Two LFA-1's on our programmed cells bind to the same ICAM dimer on a damaged cardiomyocyte. | ||
| + | |||
| + | 3. 50% of the time LFA-1 – Cub will be in close proximity of LFA-1 – NubG. | ||
| + | |||
| + | 4. When this occurs, the Cub and NubG of split ubiquitin interact and reconstitute the ubiquitin. | ||
| + | |||
| + | 5. Ubiquitin protease then cleaves the Cub portion of ubiquitin and releases the transcription factor LexA-VP16. | ||
| + | |||
| + | 6. LexA-VP16 travels to the nucleus, binds to the lexA operator minimal promoter, and effectors such as N-cadherin, VEGF, are upregulated. | ||
| + | |||
[[Image:Icam.jpg]] | [[Image:Icam.jpg]] | ||
| + | |||
| + | |||
| + | == Reasoning == | ||
| + | |||
| + | ICAM – LFA-1 Construct: | ||
| + | |||
| + | ICAM is upregulated in damaged cardiomyocytes after an insult (infarct). Many other proteins are also upregulated, myosin, CRP, etc, all were potential candidates. We then began to look at the receptors for these ligands. | ||
| + | |||
| + | LFA-1 is the ICAM receptor, it is usually found on immune cells. Normal use is for the rolling phenomena along the blood brain barrier to help cells get through. | ||
| + | |||
| + | LFA-1, is made up of CD11a (alpha-L integrin) and CD18(beta-2 integrin). Initial interest was due to believed dimerization of these parts when encountering ICAM. This is not the case, they are associated beforehand, but only become activated when bound to ICAM. | ||
| + | |||
| + | However, ICAM is a dimer when bound to the membrane (reference and picture). With this information, there was hope that there would be two receptor binding domains. This is true, allowing two LFA-1 constructs to bind to one ICAM molecule. Looked at the literature to see how close the two LFA-1 molecules might be to each other. Springer (reference), showed using FRET analysis that the two molecules are close enough to cause to get fluorescence. | ||
| + | |||
| + | Plan: attach a system to the intracellular ends of LFA-1, that turns on the expression of NCAD, allowing the cells to form better connections with the damaged hearts. The expression should only turn on when LFA-1 binds ICAM on the membrane. To get this to occur, we can use the fact that membrane-bound ICAM has two sites that can bind to LFA-1. If two LFA-1 molecules are attached, their c-terminal ends come into close proximity, as shown by Springer. We can then fuse a proximity dependent signaling system to the c-terminal end of LFA-1. We can only fuse to either CD-18 or CD-11a, not both; this insures that there will only be a signal upon two LFA-1 molecules binding to a single ICAM. | ||
| + | |||
| + | Our system of choice is the split ubiquitin system, developed in yeast. Ubiquitin is a polypeptide sequence that is recognized by ubiquitin protease. This system has been used to show whether two proteins interact. The c-terminal of ubiquitin will be fused to one protein, while having a transcription factor bound to its other end. The n-terminal of ubiquitin is fused to another protein. If they come within close proximity of each other, they will bind together and will be cleaved, releasing the transcription factor. In the yeast system, the transcription factor was used to turn on a reporter gene, such as GFP. So if two proteins interact, the cell will fluoresce. | ||
| + | |||
| + | We will then attach the Cub and NubG (a mutated version allowing for.......) to the C-terminal end of CD18, as Springer has shown that this has not hindered activity. Also while the Cub generally binds to the c-terminal end of a protein and has a transcription factor bound to its c-terminal end, NubG is generally found bound to the n-terminal end of a protein. However, it has been shown, when trying to look at the interaction between membrane bound proteins, that NubG can also bind to the c-terminal end of a protein. | ||
| + | |||
| + | To increase the likelihood of Cub and Nub being able to bind together, we put a GS-linker between CD-18 and either ubiquitin fragment. This idea was borrowed from the split intein system. The GS-linker provides flexibility and reach, which increases the likelihood of Cub and NubG interacting. | ||
| + | |||
| + | A commonly used transcription factor in Yeast is mLexA-VP16AD, the same one used in the intein system. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | 1. ICAM – upregulated in damaged heart tissue | ||
| + | 2. ICAM – Dimer when membrane bound | ||
| + | 3. Initial idea: since two LFA-1s can bind, use that as a “dimerization to release signal. | ||
| + | 4. Literature on LFA-1 binding Springer FRET analysis | ||
| + | |||
| + | |||
| + | Overview of Fixed Damaged Hearts Project: | ||
| + | Our team decided to explore the possibility of regaining function of damaged cardiac tissue using cell therapy. As it has already been shown that stem cells can help hearts regain function, our hope is to promote a sheet formation covering the entire damaged area, and ensuring that the cells form into myocardial cells. | ||
| + | In damaged tissue, CRP, myosin, ICAM, and other molecules are up regulated. We will create a synthetic signaling system that recognizes these ligands, and increases cell adhesion molecules, such as NCAD, and promotes cell differentiation/prevent apoptosis. | ||
| + | ICAM-1 is a plausible ligand due to the fact that when membrane bound it forms a dimer. This allows for specificity as soluble, monomeric, ICAM-1 will not induce a cell response. Also, having two functional binding sites for its receptor LFA-1, allows for the possibility of creating a synthetic dimerization signal. Using a synthetic signal has the advantage of avoiding cross talk with normal cell processes and signals. The general idea was that when two LFA-1 molecules come into close proximity they would release a transcription factor that would turn on our genes of interest. | ||
| + | Don’t know whether I should talk about Split Ubiquitin system now or the Springer paper now. | ||
| + | Initially we believed we would use the same split intein system as described for CRP and myosin, so before designing anything, we needed to research on whether it would be plausible to use this model. Springer et al. 2003, using LFA-1 as the model to research integrin function, showed that this was plausible. They used FRET analysis to determine the interaction between the two subunits of LFA-1, CD11a and CD18. To see if there was background fluorescence due to two LFA-1 Molecules being in close proximity to each other, yielded positive results. If the proteins are close enough to cause fluorescence, then it would be plausible that fusion proteins attached by a gslinker could interact. We decided to attach our fusion constructs to CD18 c-terminal domain because that is what the Springer Lab did when looking at background fluorescence. | ||
| + | As mentioned earlier, we did not use the split intein system for this method. The intein system requires that one half is bound to the c-terminal of a protein and the other half is bound to the n-terminal of a different protein. As we need both constructs to bind to the c-terminal domain, we switched to the split ubiquitin system, described in yeast, which has been shown to work in mammalian system. Ubiquitin, which is cleaved by ubiquitin protease, can be split in two so that it is non-functional unless the two pieces come into close proximity. Then they bind together and ubiquitin protease will cleave them. System generally works by binding mLexA-VP16AD to the c-terminal end of Cub, and both nub and cub and bound to different proteins. We will attach both Cub and Nub to CD18 with a gslinker, which allows a greater range of mobility and a higher chance of interacting with each other. When the joined ubiquitin is cleaved it will release mLexA-VP16AD, which will then travel to the nucleus, bind to the LexAoperator and transcribe our gene of interest (NCAD). | ||
| + | |||
| + | References: | ||
| + | JOSE ́ M. CASASNOVAS, THILO STEHLE, JIN-HUAN LIU, JIA-HUAI WANG, AND TIMOTHY A. SPRINGER. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. PNAS, 1998. | ||
| + | |||
| + | Minsoo Kim, Christopher V. Carman, Timothy A. Springer. Bidirectional Transmembrane Signaling by Cytoplasmic Domain Separation in Integrins. Science, 2003. | ||
| + | |||
| + | Eileen ROJO-NIERSBACH, Debra MORLEY, Stephanie HECK, and Norbert LEHMING. A new method for the selection of protein interactions in mammalian cells. Biochemistry Journal, 2000. | ||
| + | |||
| + | Malcolm S Steinberg and Patrick M McNutt. Cadherins and their connections: adhesion junctions have broader functions. Current Opinion in Cell Biology, 1999. | ||
| + | |||
| + | Patricia L. Reilly Joseph R. Woska, I. Deborah D. Jeanfavre Eugene M~Nally Robert Rothlein and Barbara-lean Bormann The Native Structure of Intercellular Adhesion Molecule-1 (ICAM- 1) Is a Dimer Correlation with Binding to LFA-1. Journal of Immunology, 1995. | ||
Latest revision as of 03:54, 27 October 2007
How it works
1. Infarced myocardium induces expression of ICAM.
2. Two LFA-1's on our programmed cells bind to the same ICAM dimer on a damaged cardiomyocyte.
3. 50% of the time LFA-1 – Cub will be in close proximity of LFA-1 – NubG.
4. When this occurs, the Cub and NubG of split ubiquitin interact and reconstitute the ubiquitin.
5. Ubiquitin protease then cleaves the Cub portion of ubiquitin and releases the transcription factor LexA-VP16.
6. LexA-VP16 travels to the nucleus, binds to the lexA operator minimal promoter, and effectors such as N-cadherin, VEGF, are upregulated.
Reasoning
ICAM – LFA-1 Construct:
ICAM is upregulated in damaged cardiomyocytes after an insult (infarct). Many other proteins are also upregulated, myosin, CRP, etc, all were potential candidates. We then began to look at the receptors for these ligands.
LFA-1 is the ICAM receptor, it is usually found on immune cells. Normal use is for the rolling phenomena along the blood brain barrier to help cells get through.
LFA-1, is made up of CD11a (alpha-L integrin) and CD18(beta-2 integrin). Initial interest was due to believed dimerization of these parts when encountering ICAM. This is not the case, they are associated beforehand, but only become activated when bound to ICAM.
However, ICAM is a dimer when bound to the membrane (reference and picture). With this information, there was hope that there would be two receptor binding domains. This is true, allowing two LFA-1 constructs to bind to one ICAM molecule. Looked at the literature to see how close the two LFA-1 molecules might be to each other. Springer (reference), showed using FRET analysis that the two molecules are close enough to cause to get fluorescence.
Plan: attach a system to the intracellular ends of LFA-1, that turns on the expression of NCAD, allowing the cells to form better connections with the damaged hearts. The expression should only turn on when LFA-1 binds ICAM on the membrane. To get this to occur, we can use the fact that membrane-bound ICAM has two sites that can bind to LFA-1. If two LFA-1 molecules are attached, their c-terminal ends come into close proximity, as shown by Springer. We can then fuse a proximity dependent signaling system to the c-terminal end of LFA-1. We can only fuse to either CD-18 or CD-11a, not both; this insures that there will only be a signal upon two LFA-1 molecules binding to a single ICAM.
Our system of choice is the split ubiquitin system, developed in yeast. Ubiquitin is a polypeptide sequence that is recognized by ubiquitin protease. This system has been used to show whether two proteins interact. The c-terminal of ubiquitin will be fused to one protein, while having a transcription factor bound to its other end. The n-terminal of ubiquitin is fused to another protein. If they come within close proximity of each other, they will bind together and will be cleaved, releasing the transcription factor. In the yeast system, the transcription factor was used to turn on a reporter gene, such as GFP. So if two proteins interact, the cell will fluoresce.
We will then attach the Cub and NubG (a mutated version allowing for.......) to the C-terminal end of CD18, as Springer has shown that this has not hindered activity. Also while the Cub generally binds to the c-terminal end of a protein and has a transcription factor bound to its c-terminal end, NubG is generally found bound to the n-terminal end of a protein. However, it has been shown, when trying to look at the interaction between membrane bound proteins, that NubG can also bind to the c-terminal end of a protein.
To increase the likelihood of Cub and Nub being able to bind together, we put a GS-linker between CD-18 and either ubiquitin fragment. This idea was borrowed from the split intein system. The GS-linker provides flexibility and reach, which increases the likelihood of Cub and NubG interacting.
A commonly used transcription factor in Yeast is mLexA-VP16AD, the same one used in the intein system.
1. ICAM – upregulated in damaged heart tissue
2. ICAM – Dimer when membrane bound
3. Initial idea: since two LFA-1s can bind, use that as a “dimerization to release signal.
4. Literature on LFA-1 binding Springer FRET analysis
Overview of Fixed Damaged Hearts Project:
Our team decided to explore the possibility of regaining function of damaged cardiac tissue using cell therapy. As it has already been shown that stem cells can help hearts regain function, our hope is to promote a sheet formation covering the entire damaged area, and ensuring that the cells form into myocardial cells.
In damaged tissue, CRP, myosin, ICAM, and other molecules are up regulated. We will create a synthetic signaling system that recognizes these ligands, and increases cell adhesion molecules, such as NCAD, and promotes cell differentiation/prevent apoptosis.
ICAM-1 is a plausible ligand due to the fact that when membrane bound it forms a dimer. This allows for specificity as soluble, monomeric, ICAM-1 will not induce a cell response. Also, having two functional binding sites for its receptor LFA-1, allows for the possibility of creating a synthetic dimerization signal. Using a synthetic signal has the advantage of avoiding cross talk with normal cell processes and signals. The general idea was that when two LFA-1 molecules come into close proximity they would release a transcription factor that would turn on our genes of interest.
Don’t know whether I should talk about Split Ubiquitin system now or the Springer paper now.
Initially we believed we would use the same split intein system as described for CRP and myosin, so before designing anything, we needed to research on whether it would be plausible to use this model. Springer et al. 2003, using LFA-1 as the model to research integrin function, showed that this was plausible. They used FRET analysis to determine the interaction between the two subunits of LFA-1, CD11a and CD18. To see if there was background fluorescence due to two LFA-1 Molecules being in close proximity to each other, yielded positive results. If the proteins are close enough to cause fluorescence, then it would be plausible that fusion proteins attached by a gslinker could interact. We decided to attach our fusion constructs to CD18 c-terminal domain because that is what the Springer Lab did when looking at background fluorescence.
As mentioned earlier, we did not use the split intein system for this method. The intein system requires that one half is bound to the c-terminal of a protein and the other half is bound to the n-terminal of a different protein. As we need both constructs to bind to the c-terminal domain, we switched to the split ubiquitin system, described in yeast, which has been shown to work in mammalian system. Ubiquitin, which is cleaved by ubiquitin protease, can be split in two so that it is non-functional unless the two pieces come into close proximity. Then they bind together and ubiquitin protease will cleave them. System generally works by binding mLexA-VP16AD to the c-terminal end of Cub, and both nub and cub and bound to different proteins. We will attach both Cub and Nub to CD18 with a gslinker, which allows a greater range of mobility and a higher chance of interacting with each other. When the joined ubiquitin is cleaved it will release mLexA-VP16AD, which will then travel to the nucleus, bind to the LexAoperator and transcribe our gene of interest (NCAD).
References: JOSE ́ M. CASASNOVAS, THILO STEHLE, JIN-HUAN LIU, JIA-HUAI WANG, AND TIMOTHY A. SPRINGER. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. PNAS, 1998.
Minsoo Kim, Christopher V. Carman, Timothy A. Springer. Bidirectional Transmembrane Signaling by Cytoplasmic Domain Separation in Integrins. Science, 2003.
Eileen ROJO-NIERSBACH, Debra MORLEY, Stephanie HECK, and Norbert LEHMING. A new method for the selection of protein interactions in mammalian cells. Biochemistry Journal, 2000.
Malcolm S Steinberg and Patrick M McNutt. Cadherins and their connections: adhesion junctions have broader functions. Current Opinion in Cell Biology, 1999.
Patricia L. Reilly Joseph R. Woska, I. Deborah D. Jeanfavre Eugene M~Nally Robert Rothlein and Barbara-lean Bormann The Native Structure of Intercellular Adhesion Molecule-1 (ICAM- 1) Is a Dimer Correlation with Binding to LFA-1. Journal of Immunology, 1995.