Imperial/Wet Lab/Results/Res1.8
From 2007.igem.org
Testing DNA concentration of pTet-GFPmut3b In vitro
Aims
To determine if the optimum concentration of [http://partsregistry.org/Part:BBa_I13522 pTet-GFPmut3b] in vitro.
Materials and Methods
Link to Protocols page
Results
Controls:
- Negative Control- Nuclease Free Water was added instead of DNA
Constants:
- Temperature - 37°C
- Total Volume - 60µl
Raw Data
Discussion
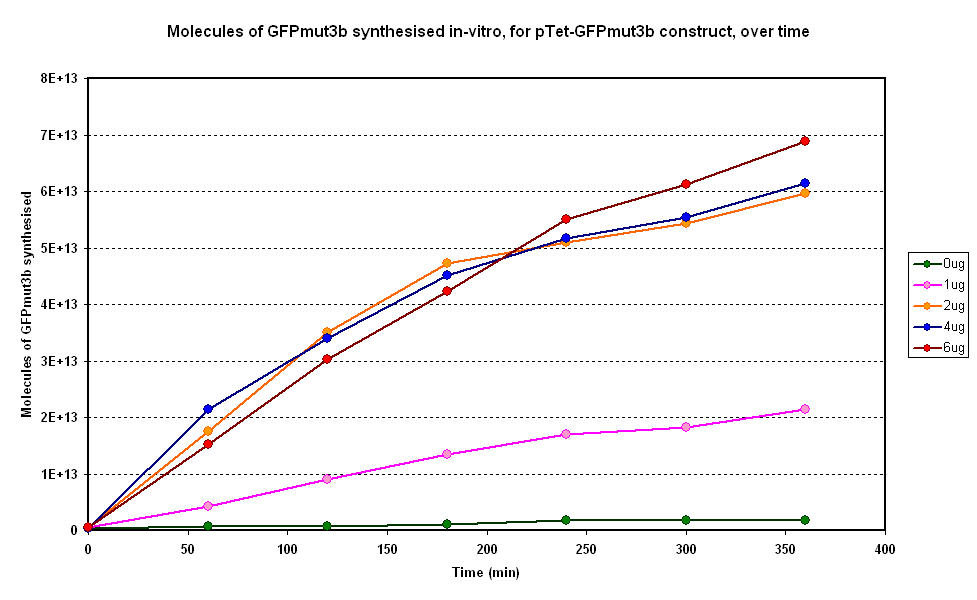
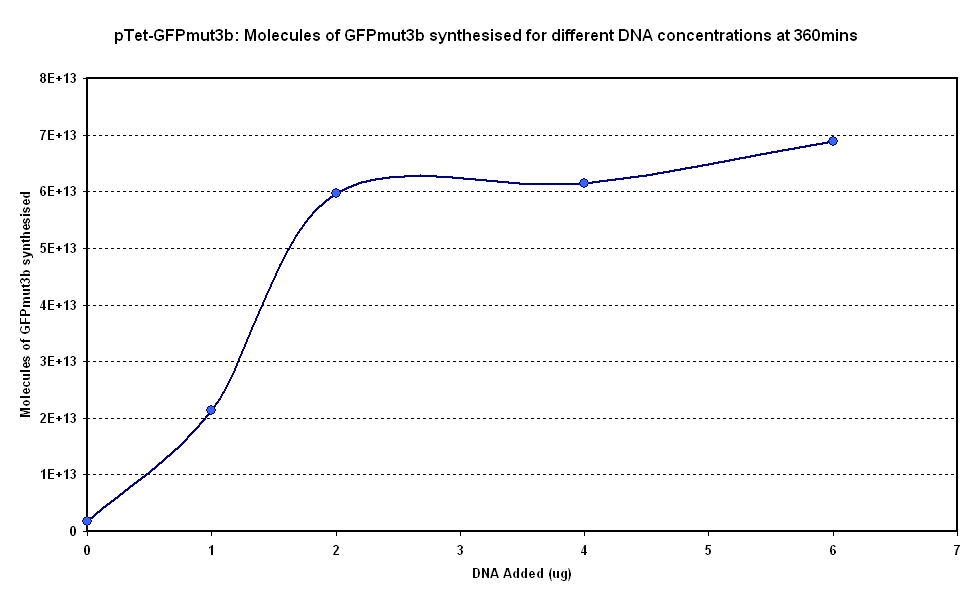
Figure 1.1 and Figure 1.2. show us that the synthesis of GFPmut3b increases with DNA added. Though the molecules of GFPmut3b synthesised increases with increasing DNA concentration, even after a concentration of 4µg. This is not what was expected because of the nature of the cell free chassis which is optimum DNA concentration = 4µg. But the increase in fluorescent moecules synthesised by increasing DNA concentration is not very big. If DNA concentration is increased from 2µg to 6µg (a percentage increase of 200%), it reuslts in only about 15% increase (at 360min) in GFPmut3b molecules synthesised. Thus it was decided that a DNA concentration of 2 or 4&micor;g will be used for testing pTet-GFPmut3b construct.
Conclusion
To conclude the following approximations can be made:
Optimum DNA concentrations - 2 or 4µg of DNA