Michigan
From 2007.igem.org
For more information and documentation, please visit our [http://synthbio.engin.umich.edu/wiki/index.php/Main_Page wiki!]
General Project Description
We have designed a Divide-By-Two (DBT) circuit, extensively modeled the system to predict behavior, and fabricated the device. We have also designed a landing pad compatible with all BioBricks in order to increase the stability of the DBT device, due to the noisy behavior predicted from our modeling.
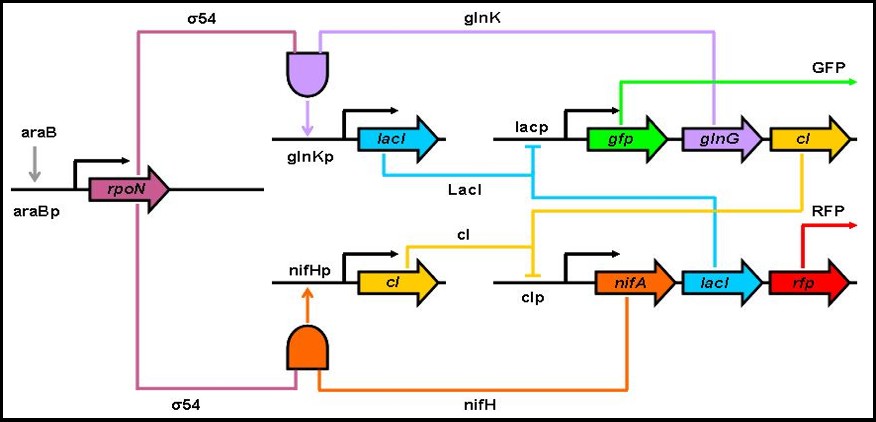
Divide-By-Two Circuit
The DBT circuit is a single-input toggle switch with the green and red fluorescent proteins (GFP and RFP for short) to monitor product levels. If the system is at a steady state, with high concentrations of GFP and low RFP levels, adding arabinose will cause a switch in the protein levels. As a result, another steady state will be reached in which RFP is at a high concentration and there are small GFP levels. Adding arabinose again causes a switch back to the original steady state of the system (GFP at high levels, RFP at low ones). The crucial elements of the circuit are the two AND-gates, which require two specific molecules in order to activate transcription of the gene driven by the specific promoter. The specific mechanics of the DBT circuit are as follows: If we start at a state where the levels of GFP, glnK, cI are high and the levels of nifA, nifH, RFP, and the sigma-54 factor are low, by adding arabinose, we increase the levels of sigma-54. The high levels of sigma-54 and glnK causes the lacI gene to be transcribed by “activating” the glnKp promoter. lacI inhibits further production of GFP, glnK, and cI. At the same time, because of the low levels of nifH, cI will not be transcribed as much as lacI. By the time the arabinose is used up, the levels of GFP, glnG, and cI will have decreased substantially and the levels of nifA, lacI, and RFP will have increased. Similar behavior to the one described above will occur if arabinose is added again.
|
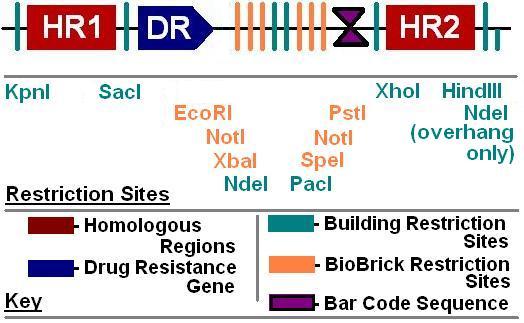
BioBrick Landing Pad
The goal of this project is to develop tools to aid synthetic biologists in inserting genes and other DNA sequences onto the E. coli chromosome. While a few “simple” landing pads are already available for placement of DNA onto the chromosome, our project seeks to develop a general strategy for constructing landing pads that are exceptionally easy to place onto the chromosome and have advanced features. Among these advanced features are limiting noise from external promoters, allowing simplified phenotype screening and genetic analysis, allowing BioBrick-compatability, and allowing sequential insertion of DNA modules into the same chromosomal locus, where they will form a set of nested sequences.
Check out these wiki pages:
|
|---|
Meet our team!
Students
|
Advisors
|
|---|