NYMU Taipei/Lab Notes/2007 10 4
From 2007.igem.org
(Difference between revisions)
(→Re-check the concentration of inserts and vectors) |
(→ligation setup) |
||
| Line 34: | Line 34: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | == ligation setup== | + | == ligation setup == |
| + | * ligation I: pCinRHSL (vector) + OmpRBS (insert) | ||
| + | <table border=1> | ||
| + | <tr><td>pCinRHSL (vector)</td><td>3 uL</td></tr> | ||
| + | <tr><td>OmpRBS (insert)</td><td>14 uL</td></tr> | ||
| + | <tr><td>10X buffer</td><td>2 uL</td></tr> | ||
| + | <tr><td>T4 ligase</td><td>1 uL</td></tr> | ||
| + | </table> | ||
Revision as of 14:48, 4 October 2007
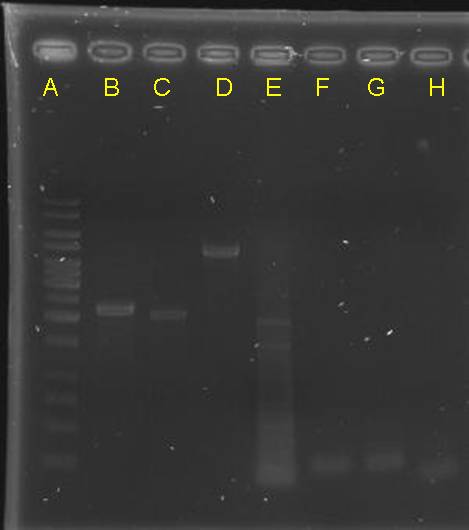
digestion check of CinR+HSL+D-term,pCinRHSL and pOmpC (vectors)
 |
|
- after gel separation, their O.D. was checked. However, it is too low
Re-check the concentration of inserts and vectors
ligation setup
- ligation I: pCinRHSL (vector) + OmpRBS (insert)
| pCinRHSL (vector) | 3 uL |
| OmpRBS (insert) | 14 uL |
| 10X buffer | 2 uL |
| T4 ligase | 1 uL |