Nhu Nguyen Notebook
From 2007.igem.org
| Line 70: | Line 70: | ||
I checked on my PCR tubes today and found out that the PCR tubes that contain phusion did not work. The PCR tubes that expand worked. This time, ThpA came out right; however, ThpB did not come correctly. <br> | I checked on my PCR tubes today and found out that the PCR tubes that contain phusion did not work. The PCR tubes that expand worked. This time, ThpA came out right; however, ThpB did not come correctly. <br> | ||
<br> | <br> | ||
| - | After realizing that I put in the wrong primers for ThpB, I | + | After realizing that I put in the wrong primers for ThpB, I put ThpB in the PCR once more and the result was satisfying. The first PCR tube for ThpB needed to have ThpBF and ThpBIR. The second PCR tube need to have ThpBR and ThpBIF. After I took out my PCR tubes, I gelled it to make sure that it has the right ban and then I cleaned up.<br> |
<br> | <br> | ||
I also cleaned up ThpA and ThpC and digested them. After putting them in the incubator for 2 hours, I took them out and cleaned them up again. After digestion, I underwent ligation by adding vector 9145, my insert (ThpA and ThpC), ligase buffer, ligase and water. After that, I let the tubes sit for 30 minutes to an hour.<br> | I also cleaned up ThpA and ThpC and digested them. After putting them in the incubator for 2 hours, I took them out and cleaned them up again. After digestion, I underwent ligation by adding vector 9145, my insert (ThpA and ThpC), ligase buffer, ligase and water. After that, I let the tubes sit for 30 minutes to an hour.<br> | ||
| Line 76: | Line 76: | ||
<br> | <br> | ||
After ligation was finished, I carried out transformation. I needed 220uL of competent cells and salt solution ( 30uL of KCM and 50uL H2O). After added the salt solution to the competent cells, I added 100uL of the solution to each ligation tube. I mixed it and put them on ice for 10 minutes. After that, I heat shock if for 90 seconds in 42C. I put it back on ice for 2 minutes. I added 110uL to an agar dish, and I used the beads to spread the solution. In the end, I put the agar dishes in the incubator for 16 hours.<br> | After ligation was finished, I carried out transformation. I needed 220uL of competent cells and salt solution ( 30uL of KCM and 50uL H2O). After added the salt solution to the competent cells, I added 100uL of the solution to each ligation tube. I mixed it and put them on ice for 10 minutes. After that, I heat shock if for 90 seconds in 42C. I put it back on ice for 2 minutes. I added 110uL to an agar dish, and I used the beads to spread the solution. In the end, I put the agar dishes in the incubator for 16 hours.<br> | ||
| + | <br> | ||
| + | The correct reverse primer for ThpD came and I added water to the tube. After vortexing it, I took out 1uL of primer and added to a 9uL of water which makes a total of 10uL. After adding the correct primers, template, enzyme, buffer, dNtps, and water, I placed my PCR tubes into the PCR machine. Unfortunately, PCR did not work for ThpD. | ||
| + | |||
| + | |||
| + | '''[[User:Nnguyen|Nnguyen]] 19:21, 29 June 2007 (EDT)''' | ||
| + | ---- | ||
| + | I came in today hoping that I can get ThpD to work. I tried three different techniques hoping that PCR would work. The first PCR tube, I used the enzyme phusion and HF buffer 5x and everything else is the same as the previous PCR tubes. The second PCR tube, I used the enzyme phusion and GC buffer 5x. The third PCR tube, I used the enzyme expand and expand buffer 10x. After taking the tubes out of the PCR and gelling it, the tube with expand did not have a band; the tube with phusion and GC buffer had too many bands, and the tube with phusion and HF buffer showed a satisfying ban. <br> | ||
<br> | <br> | ||
Revision as of 23:21, 29 June 2007
Nnguyen 15:40, 25 June 2007 (EDT)
Today I came in and I started my tutorial on internal sites because I missed a week of work last week. I completed the tutorial, and I was given my task for the week. Chris gave me a template to work on. The gene is called Streptomyces chrysomallus. There are four genes of interest in the template, and I had to make oligos for these genes. I had to construct two oligos for each gene, a forward primer and a reverse primer. In order to construct the oligos, I used the enzymes BglII and XhoI. BglII was used for the forward primer and XhoI was used for the reverse primer. In 2 out 4 genes, there were internal sites which meant that I had to make additional oligos.For gene ThpA, EcoRI was within it and so I had to make a primer that would remove it. The sequence for EcoRI was gaattc, and I changed it to gagttc. gag and gaa both code for the same amino acid which is glutamic acid. XhoI was within gene ThpB, and again I had to make a primver that would remove the restriction enzyme.
The sequence for XhoI is ctcgag, and I changed it to cttgag. ctc and ctt both code for the same amino acid which is leucine. In order to remove XhoI from ThpB, I construct a forward primer and a reverse primer. The reverse primer is the complimentary side of the forward primer. Compared to the oligos that I constructed for the beginning and end of each gene, the primers used to remove XhoI do not have restriction enzymes on them. Compared to ThpB, ThpA only has 2 primers and not 4 primers because the EcoRI was very close to the beginning of the gene. Therefore, I incorporated EcoRI into the forward primer for the gene ThpA. In the end, I ended up with 10 oligos.
At the end of the day, Sam showed the high school students how to pour agar into plates, and we left the plates on the counter throughout the night.
Nnguyen 15:40, 26 June 2007 (EDT)
I came into work today and helped the other high school students a little bit with their project while I was waiting for my oligos to arrive. They have not arrived yet; however, in the meantime, Sam and Arthur showed us how to make LB Agar and LB Broth. We poured 500mL of water into each glass bottle. For LB Agar, we added 20g of agar powder, and for LB Broth, we added 12.5g of broth powder. After adding the right amount into the 500mL of water, we shook the bottle until the powder dissolved. After that, we put all the glass bottles in a tray, untwisted the cap so that when they're put in the machine, they wouldn't break due to high pressure, and we added water and tape the caps. After we finished with the preparations, we put the tray into the autoclave machine for 25 minutes; however, it is better to let it sit in there for awhile to let it cool down. The agar that was placed in the autoclave machine did not work because the previous person who used it did not reset. We took out the agar bottles and let the machine exhaust for a while, and then we put it back in the autoclave for 25 minutes. After it finished, we let it stayed in there for a while so it can cool down. After taking them out, we added antibiotics to the agar bottles. We added AMP (antibiotics) and CAM (antibiotics) to 3 of the agar bottles, and we shook it. In the other agar bottles, we only added AMP. After shaking the bottles, we waited until the bubbles disappear and then we started pouring them into plates. If there were bubbles left in the plate, we put the flame directly on the agar so the bubbles go away. The plates that consist of AMP, we marked it with the color red. The plates that consist of AMP and CAM, we marked it with blue and red.
Nnguyen 23:57, 26 June 2007 (EDT)
At around 2:00, the oligos that I ordered yesterday arrived. I added water to each tube because the primers came in a form of powder. On each tube, it tells how much powder it is in nanomol. I multiplied the number of nanomol by 10 and that was how much water I added. For example, I added 385 microL of water into nn001.
nn001: 38.5 nmol
nn002: 27.5 nmol
nn003: 29.0 nmol
nn004: 29.4 nmol
nn005: 37.7 nmol
nn006: 33.6 nmol
nn007: 26.5 nmol
nn008: 29.9 nmol
nn009: 27.5 nmol
nn010: 31.3 nmol
The DNA sequences for each of primers are:
nn001:cgtacAGATCTatgaccgccgcaccagcagattttgcccgtgcccgaagcgaGttcctcagtatcg
nn002:cgcatctcgagtcaggtggcgaacgggccta
nn003:ctagaAGATCTgtgaccatcaccccgcccgc
nn004:cgtcactcgagtcaggccgtttcgcggacgc
nn005:ggttcgagcggctcctTgaggaccagggctccggac
nn006:gtccggagccctggtcctcAaggagccgctcgaacc
nn007:ctagtAGATCTgtgatcgtccgatcgttcag
nn008:gtctactcgagtcaggcctcctcggtcagca
nn009:cggatAGATCTatgaccaccgaagtacgcgc
nn010:gtctactcgagtcaggcctcctcggtcagca
After adding water, I vortex them so that all the loose particles would settle down, and the tubes were put on ice. After placing the tubes in ice, I started predicting my PCR product. My predictions are: ThpA=553 bp; ThpB=1285 bp; ThpC=421 bp; ThpD=916 bp.At the end of the day, I put all 10 tubes in the freezer (below 20).
Nnguyen 14:06, 27 June 2007 (EDT)
Today, I came in and found out that my template has not yet arrived. While I was updating my notebook, I realized that I have the same two primers (nn008 and nn010). nn008 is correct; however, nn010 is incorrect. I just finished putting my correct primer (nn011) on order.
The correct DNA sequence for nn010 is:
nn011:gtcagctcgagctacttcaccggggtgaagt
Without nn011, I am still able to continue with my work even if my template arrives today.
When my template arrived, the DNA was embedded in an organism and so I had to isolate the DNA. I used the Ultra Clean Mircobial DNA isolation Kit to isolate the DNA. After isolating the DNA i was ready for PCR. First, I had to dilute the primer and so I added 1uL of primer to a new tube and I added 9uL of water to each tube. After making PCR tubes and putting the right amount of primers (forward, reverse), template, enzyme, buffer, dNtps and water, I put my PCR tubes in the PCR machine. I used the 3-step program and the process took 1 hour and 14 minutes.
After PCR #1 was done, I ran it through the gel to see whether if my predictions were right. Unfortunately, ThpA and ThpB did not turn out right, ThpC was correct.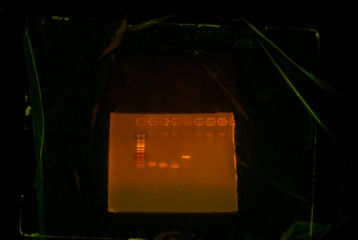
ThpA (2nd well) and ThpB (3nd and 4rd well) did not work and ThpC (5th well) worked.
I attempted to PCR again. I had 6 PCR tubes: 3 PCR tubes containing the enzyme expand and Dmso 100% and the other 3 PCR tubes contained the enzyme phusion and no Dmso 100%.
Nnguyen 16:14, 28 June 2007 (EDT)
I checked on my PCR tubes today and found out that the PCR tubes that contain phusion did not work. The PCR tubes that expand worked. This time, ThpA came out right; however, ThpB did not come correctly.
After realizing that I put in the wrong primers for ThpB, I put ThpB in the PCR once more and the result was satisfying. The first PCR tube for ThpB needed to have ThpBF and ThpBIR. The second PCR tube need to have ThpBR and ThpBIF. After I took out my PCR tubes, I gelled it to make sure that it has the right ban and then I cleaned up.
I also cleaned up ThpA and ThpC and digested them. After putting them in the incubator for 2 hours, I took them out and cleaned them up again. After digestion, I underwent ligation by adding vector 9145, my insert (ThpA and ThpC), ligase buffer, ligase and water. After that, I let the tubes sit for 30 minutes to an hour.
Note: Ligation is occuring in two different tubes (ThpA and ThpC)
After ligation was finished, I carried out transformation. I needed 220uL of competent cells and salt solution ( 30uL of KCM and 50uL H2O). After added the salt solution to the competent cells, I added 100uL of the solution to each ligation tube. I mixed it and put them on ice for 10 minutes. After that, I heat shock if for 90 seconds in 42C. I put it back on ice for 2 minutes. I added 110uL to an agar dish, and I used the beads to spread the solution. In the end, I put the agar dishes in the incubator for 16 hours.
The correct reverse primer for ThpD came and I added water to the tube. After vortexing it, I took out 1uL of primer and added to a 9uL of water which makes a total of 10uL. After adding the correct primers, template, enzyme, buffer, dNtps, and water, I placed my PCR tubes into the PCR machine. Unfortunately, PCR did not work for ThpD.
Nnguyen 19:21, 29 June 2007 (EDT)
I came in today hoping that I can get ThpD to work. I tried three different techniques hoping that PCR would work. The first PCR tube, I used the enzyme phusion and HF buffer 5x and everything else is the same as the previous PCR tubes. The second PCR tube, I used the enzyme phusion and GC buffer 5x. The third PCR tube, I used the enzyme expand and expand buffer 10x. After taking the tubes out of the PCR and gelling it, the tube with expand did not have a band; the tube with phusion and GC buffer had too many bands, and the tube with phusion and HF buffer showed a satisfying ban.