PennState/Project/Diauxie/DiauxieElimination
From 2007.igem.org
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | '''Test for Diauxie Elimination''' | ||
| + | |||
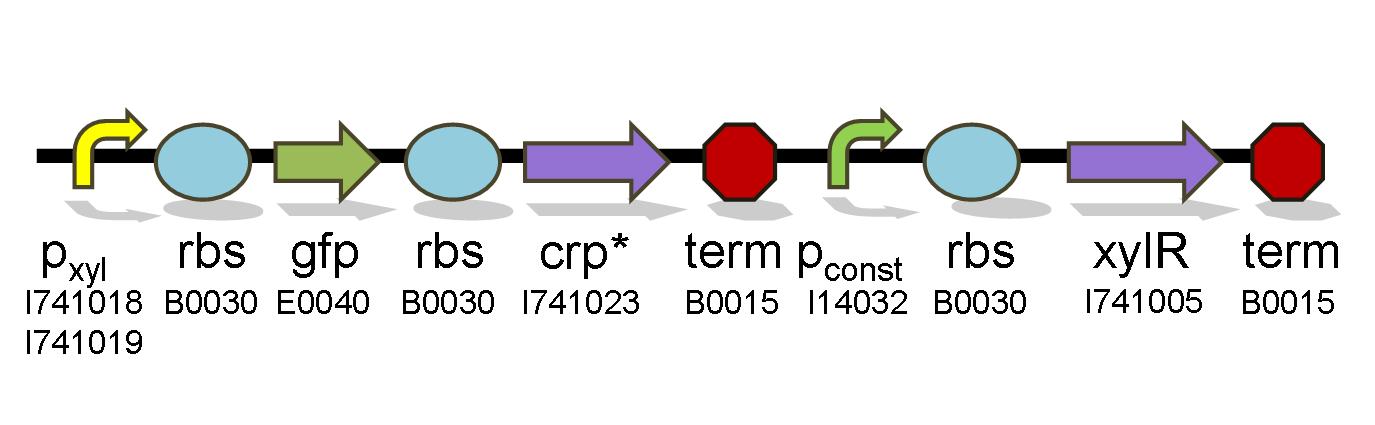
| + | To test for diauxie elimination, constitutively expressed xylR transcriptional regulator is added following the promoter test constructs. The increased level of xylR and CRP* in the cell should activate the natural chromosomal promoter for xylR, thus causing xylose metabolization even in the presence of glucose, but necessarily in the presence of xylose. | ||
| + | |||
| + | <html><img src="http://openwetware.org/images/e/e6/Dtest.jpg" id="logo" name="logo" height="150" width="550" alt="" /></html> | ||
| + | |||
| + | |||
| + | A possible model for cells naturally growing and cells growing with diauxie elimination is shown below. Elimination of diauxie with the use of CRP* could require too much of a time delay before reaching maximum growth. In this case the promoter region would need to be altered to resemble the consensus promoter sequence while keeping the xylR binding site to maintain xylose dependence. | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/2007/thumb/f/fa/Psuigem07curve2.jpg/633px-Psuigem07curve2.jpg" id="logo" name="logo" height="350" width="375" alt="" /></a> | ||
| + | |||
| + | </html> | ||
| + | |||
<html> | <html> | ||
<!-- START OF LUC'S CODE --> | <!-- START OF LUC'S CODE --> | ||
| Line 150: | Line 165: | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
<!-- END OF LUC'S CODE --> | <!-- END OF LUC'S CODE --> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</html> | </html> | ||
Latest revision as of 23:48, 26 October 2007
Test for Diauxie Elimination
To test for diauxie elimination, constitutively expressed xylR transcriptional regulator is added following the promoter test constructs. The increased level of xylR and CRP* in the cell should activate the natural chromosomal promoter for xylR, thus causing xylose metabolization even in the presence of glucose, but necessarily in the presence of xylose.
A possible model for cells naturally growing and cells growing with diauxie elimination is shown below. Elimination of diauxie with the use of CRP* could require too much of a time delay before reaching maximum growth. In this case the promoter region would need to be altered to resemble the consensus promoter sequence while keeping the xylR binding site to maintain xylose dependence.
