Purdue
From 2007.igem.org
(→Undergraduate Students) |
(→Useful References) |
||
| (33 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | == | + | == Abstract == |
| - | + | This year's Purdue 2007 iGEM team will be presenting a project called "Bacteria | |
| + | Warfare" for this year's competition hosted by MIT in November. The team is | ||
| + | composed of a group of undergraduates from different majors including Biomedical | ||
| + | Engineering, Agricultural and Biological Engineering, Biochemistry, and | ||
| + | Chemistry. Escherichia coli (E. coli) is used to conduct the microbial warfare. The design is to use two different | ||
| + | types of transformed E. coli. One type of E. coli produces protein that triggers the death gene of its opponent (but not for itself) which expresses the production of a protein toxin. One expresses green fluorescence protein and the other expresses red fluorescence protein so that the progression of the war | ||
| + | between types can be easily viewed and monitered. The team decided the general topic of a bacteria wafare at the end of spring 07 and started design and construction of the wafare in summer 07. | ||
| + | |||
| + | [[Image:Bacteria warfare.jpg]] | ||
==Contact List== | ==Contact List== | ||
| Line 11: | Line 19: | ||
=== Graduate Advisors: === | === Graduate Advisors: === | ||
*[[Purdue: John Schumm |John Schumm jschumm@purdue.edu]] <br> | *[[Purdue: John Schumm |John Schumm jschumm@purdue.edu]] <br> | ||
| + | *Alex Dimauro adimauro@purdue.edu | ||
| + | |||
=== Undergraduate Students === | === Undergraduate Students === | ||
| - | *[[Purdue: Aaron Young |Aaron Young ajyoung@purdue.edu]] <br> | + | *[[Purdue: Aaron Young |Aaron Young, ajyoung@purdue.edu]] junior biomedical engineering<br> |
| - | *[[Purdue: Meng Zhou |Meng Zhou zhoum@purdue.edu]] <br> | + | *[[Purdue: Meng Zhou |Meng Zhou, zhoum@purdue.edu]] senior chemistry <br> |
| - | *[[Purdue: Janie Stine |Janie Stine mstine@purdue.edu]] <br> | + | *[[Purdue: Janie Stine |Janie Stine, mstine@purdue.edu]] sophomore biological and food processing engineering<br> |
| - | *[[Purdue: Dan Sheik |Dan Sheik dsheik@purdue.edu]] <br> | + | *[[Purdue: Dan Sheik |Dan Sheik, dsheik@purdue.edu]] senior biochemistry<br> |
| + | * Craig Barcus, junior agricultural and biological engineering | ||
| + | |||
| + | == Useful References == | ||
| + | |||
| + | [http://intl-mmbr.asm.org/cgi/reprint/64/3/503 Programmed Cell Death in Bacteria] | ||
| + | |||
| + | [http://www.springerlink.com/content/n88p472061740k32/fulltext.pdf Spiteful Bacteria ... Who wins?] | ||
| + | |||
| + | [http://jb.asm.org/cgi/reprint/94/4/1093 colicin tolerant E coli mutants] | ||
| + | |||
| + | [http://www.nature.com/nature/journal/v428/n6981/pdf/nature02429.pdf Bacterial Rock Paper Scissors] | ||
| + | |||
| + | [http://www.che.caltech.edu/groups/fha/circuits_quorum.html Synthetic Ecosystem] | ||
== Safety== | == Safety== | ||
| - | + | Safety training was provided to all iGem team members except those already trained from lab work or from courses | |
| - | + | ||
| - | |||
Work in Lab and should have already compeleted safety training: | Work in Lab and should have already compeleted safety training: | ||
* Meng Zhou | * Meng Zhou | ||
| Line 30: | Line 51: | ||
== Project Description and Details == | == Project Description and Details == | ||
| + | |||
| + | The plan is to transform two types of E. Coli: one with plasmid 1 and the other plasmid 2. | ||
| + | |||
| + | The gene map can be find from IGEM parts: BBa_I738009 (for plasmid 1) and BBa_I738010 (for plasmid 2) | ||
| + | |||
| + | The two devices (parts assembled within a plasmid) are (in the actual DNA sequence): | ||
| + | |||
| + | [Green fluorescent protein production][AraC production][Cl regulator][ccd_B death gene] | ||
| + | |||
| + | & | ||
| + | |||
| + | [Red fluorescent protein production][Cl production][AraC regulator][ccd_B death gene] | ||
| + | |||
| + | |||
| + | |||
| + | Assembling the parts to each of the two plasmids were not finished. | ||
| + | |||
| + | -Parts DNA were amplified by introducing them to E. coli. | ||
| + | |||
| + | -Standard assembly methods were applied to constructing the devices: | ||
| + | http://partsregistry.org/Assembly:Standard_assembly | ||
| + | |||
| + | -double digestion information was found from New England Biolabs website: | ||
| + | http://www.neb.com/nebecomm/DoubleDigestCalculator.asp? | ||
| + | |||
| + | -modeling of the bacteria warfare is done and problems associated with such a design is explored. | ||
| + | |||
| + | -in the future we may continue this project if the next generation of members are interested in it. It's preferred to start the design work in spring and start the experiment and modeling in the summer to. | ||
| + | |||
| + | [[Image:Slide4.JPG]] | ||
| + | [[Image:Slide5.JPG]] | ||
| + | [[Image:Slide6.JPG]] | ||
| + | [[Image:Slide7.JPG]] | ||
| + | [[Image:Slide8.JPG]] | ||
| + | [[Image:Slide9.JPG]] | ||
| + | [[Image:Slide10.JPG]] | ||
| + | [[Image:Slide11.JPG]] | ||
| + | [[Image:Slide12.JPG]] | ||
| + | [[Image:Slide13.JPG]] | ||
| + | [[Image:Slide14.JPG]] | ||
| + | [[Image:Slide15.JPG]] | ||
| + | [[Image:Slide16.JPG]] | ||
== Photo Area == | == Photo Area == | ||
Our home building on Campus : | Our home building on Campus : | ||
| - | + | [[Image:Bindly_for_ppt.jpg]] | |
| - | [ | + | |
Latest revision as of 15:08, 26 October 2007
Contents |
Abstract
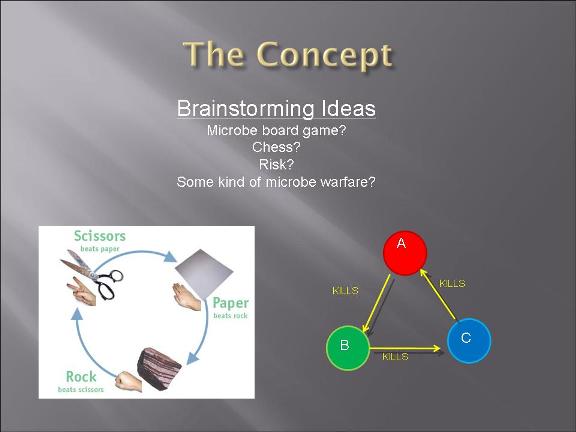
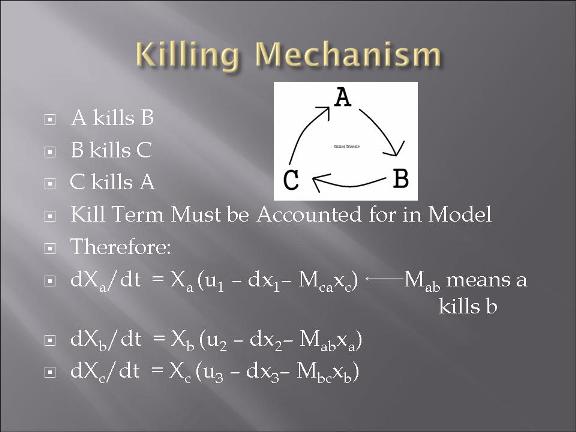
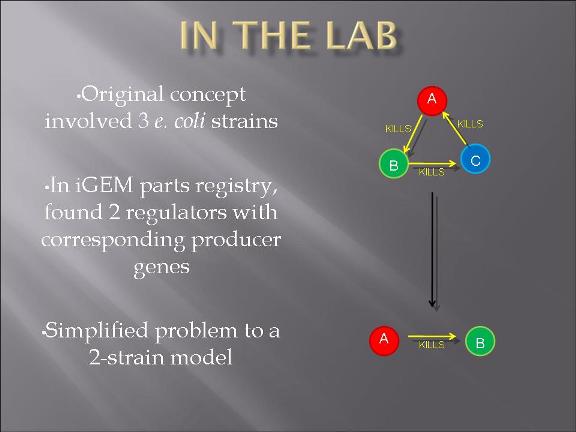
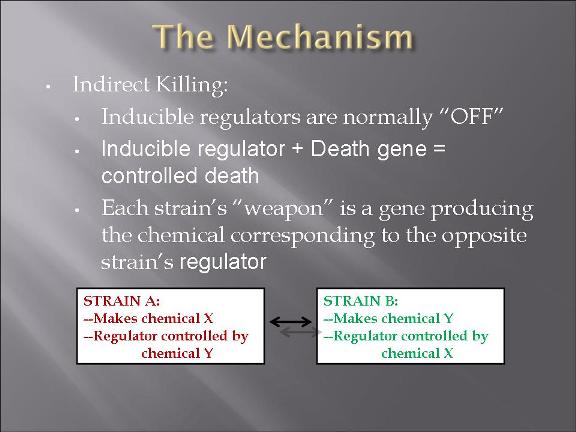
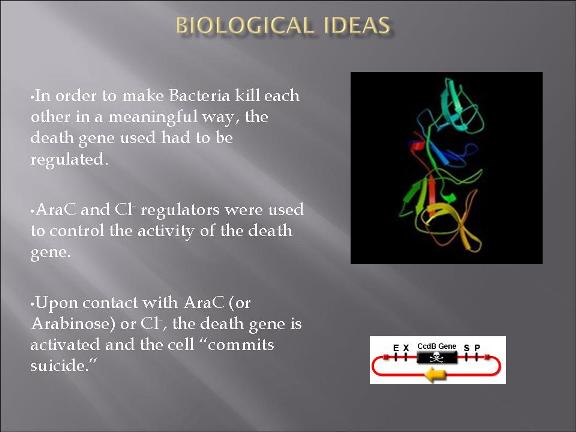
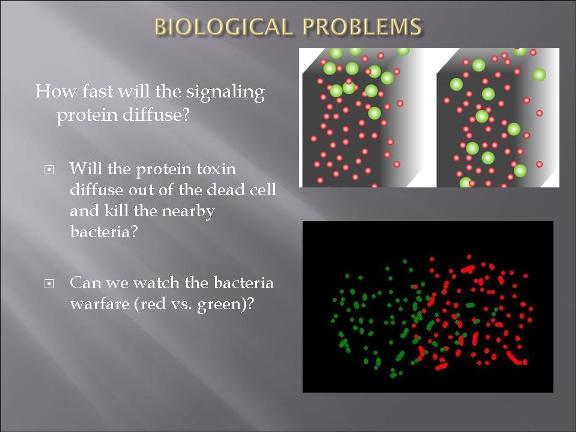
This year's Purdue 2007 iGEM team will be presenting a project called "Bacteria Warfare" for this year's competition hosted by MIT in November. The team is composed of a group of undergraduates from different majors including Biomedical Engineering, Agricultural and Biological Engineering, Biochemistry, and Chemistry. Escherichia coli (E. coli) is used to conduct the microbial warfare. The design is to use two different types of transformed E. coli. One type of E. coli produces protein that triggers the death gene of its opponent (but not for itself) which expresses the production of a protein toxin. One expresses green fluorescence protein and the other expresses red fluorescence protein so that the progression of the war between types can be easily viewed and monitered. The team decided the general topic of a bacteria wafare at the end of spring 07 and started design and construction of the wafare in summer 07.
Contact List
Faculty Advisors
- Dr. Rickus rickus@purdue.edu
- Dr. Clase
- Dr. Mosier
Graduate Advisors:
- John Schumm jschumm@purdue.edu
- Alex Dimauro adimauro@purdue.edu
Undergraduate Students
- Aaron Young, ajyoung@purdue.edu junior biomedical engineering
- Meng Zhou, zhoum@purdue.edu senior chemistry
- Janie Stine, mstine@purdue.edu sophomore biological and food processing engineering
- Dan Sheik, dsheik@purdue.edu senior biochemistry
- Craig Barcus, junior agricultural and biological engineering
Useful References
[http://intl-mmbr.asm.org/cgi/reprint/64/3/503 Programmed Cell Death in Bacteria]
[http://www.springerlink.com/content/n88p472061740k32/fulltext.pdf Spiteful Bacteria ... Who wins?]
[http://jb.asm.org/cgi/reprint/94/4/1093 colicin tolerant E coli mutants]
[http://www.nature.com/nature/journal/v428/n6981/pdf/nature02429.pdf Bacterial Rock Paper Scissors]
[http://www.che.caltech.edu/groups/fha/circuits_quorum.html Synthetic Ecosystem]
Safety
Safety training was provided to all iGem team members except those already trained from lab work or from courses
Work in Lab and should have already compeleted safety training:
- Meng Zhou
- Dan Sheik
Bindley Safety
Finally, for safety information specific to the labs in Bindley see the [http://openwetware.org/wiki/Rickus_Lab_Safety Rickus Lab Safety] page on openwetware
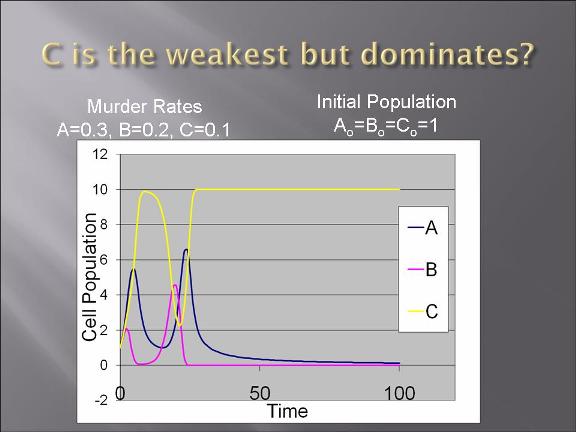
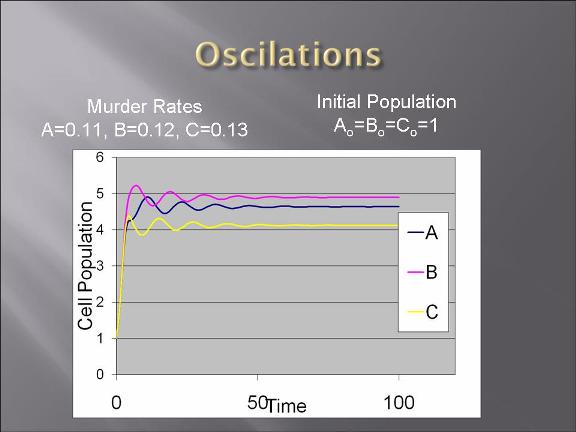
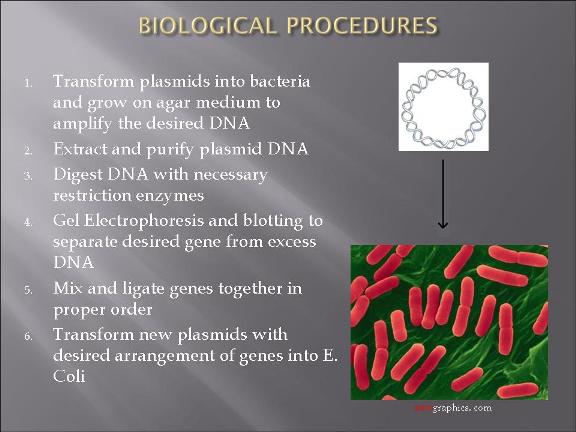
Project Description and Details
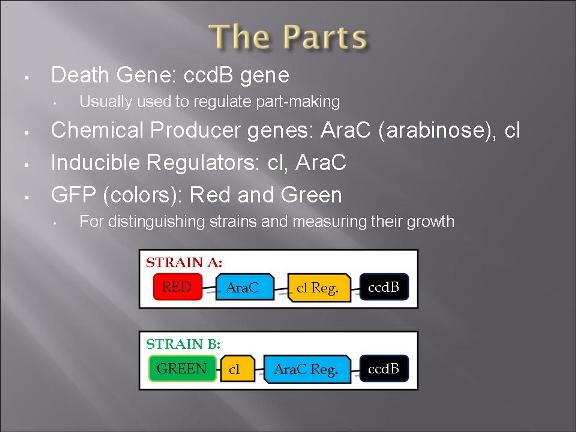
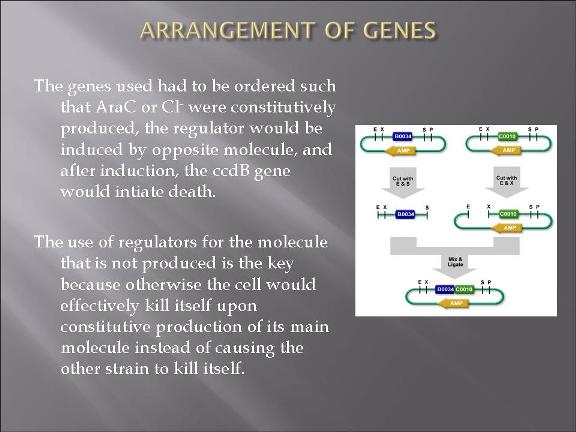
The plan is to transform two types of E. Coli: one with plasmid 1 and the other plasmid 2.
The gene map can be find from IGEM parts: BBa_I738009 (for plasmid 1) and BBa_I738010 (for plasmid 2)
The two devices (parts assembled within a plasmid) are (in the actual DNA sequence):
[Green fluorescent protein production][AraC production][Cl regulator][ccd_B death gene]
&
[Red fluorescent protein production][Cl production][AraC regulator][ccd_B death gene]
Assembling the parts to each of the two plasmids were not finished.
-Parts DNA were amplified by introducing them to E. coli.
-Standard assembly methods were applied to constructing the devices: http://partsregistry.org/Assembly:Standard_assembly
-double digestion information was found from New England Biolabs website: http://www.neb.com/nebecomm/DoubleDigestCalculator.asp?
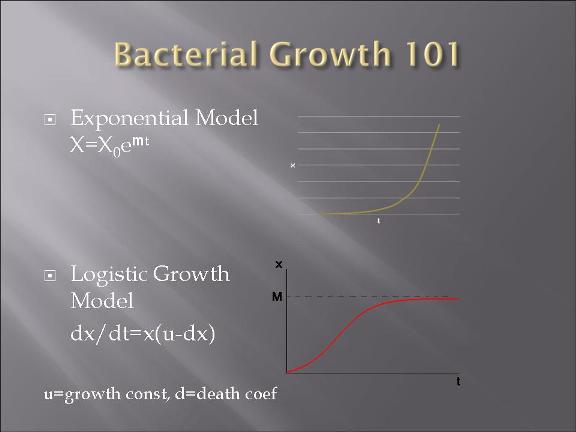
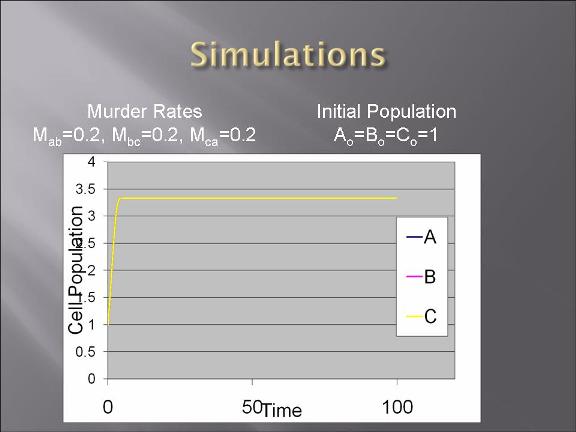
-modeling of the bacteria warfare is done and problems associated with such a design is explored.
-in the future we may continue this project if the next generation of members are interested in it. It's preferred to start the design work in spring and start the experiment and modeling in the summer to.