Samantha Liang Notebook2
From 2007.igem.org
| Line 11: | Line 11: | ||
---- | ---- | ||
'''To do:''' | '''To do:''' | ||
| - | |||
| - | |||
| - | |||
*Transform the pbad.1106A.barnase (408C) - should have a -80 of it - with RFP on a pSB3C6 plasmid (pSB3C6-J01022 from last year? check that) - will then induce with arabinose and look at it under a microscope | *Transform the pbad.1106A.barnase (408C) - should have a -80 of it - with RFP on a pSB3C6 plasmid (pSB3C6-J01022 from last year? check that) - will then induce with arabinose and look at it under a microscope | ||
*If the 1106H.Cre things are good then I'm going to have to test to see if it recombines with the genomic GenR thing | *If the 1106H.Cre things are good then I'm going to have to test to see if it recombines with the genomic GenR thing | ||
| Line 20: | Line 17: | ||
'''For tomorrow:''' <br> | '''For tomorrow:''' <br> | ||
| - | * | + | *Small scale competent cell prep of 462, 452, and 463 and transform pSB3C6-J01022 into it! - rescue and plate |
| + | *Genomic prep of 477x475 - PCR with upp locus oligos | ||
| + | *Microscopy on J01022x408C cultures | ||
| + | *If you have time, run gel on digestions, purify, ligate and transform to make I716482 pbad.1106H.Cre | ||
| + | |||
| + | |||
| + | ==[[User:Samanthaliang|Samanthaliang]] 20:22, 2 October 2007 (EDT)== | ||
*Analyze sequencing | *Analyze sequencing | ||
| - | *Grow up colonies from the | + | *Grow up colonies from the Cre transformations in Carb |
| - | *Collect Tecan data from upstairs - see if there's any visible phenotypes | + | *Grow up J01022x408C colonies with and without arabinose |
| + | *Collect Tecan data from upstairs - see if there's any visible phenotypes - BamHI and barnase hits! | ||
| + | *Grow up 462, 452, and 463 for a small scale competent cell prep (ask someone to seed them for you tomorrow!) | ||
| + | *Start digestions to make pbad.1106H.Cre | ||
Revision as of 00:22, 3 October 2007
My Construction Files
My Sequencing Files
My Biobricks
My -80 Stocks
First Notebook (June - July 19, 2007)
To do:
- Transform the pbad.1106A.barnase (408C) - should have a -80 of it - with RFP on a pSB3C6 plasmid (pSB3C6-J01022 from last year? check that) - will then induce with arabinose and look at it under a microscope
- If the 1106H.Cre things are good then I'm going to have to test to see if it recombines with the genomic GenR thing
- If the pcon.rbs.atg.lox.TT.lox.toxin things are good, then I'm going to have to get ready to knock them into the genome - need to talk to Chris for protocol but it's like a Dotsanko-Wagner thing.
For tomorrow:
- Small scale competent cell prep of 462, 452, and 463 and transform pSB3C6-J01022 into it! - rescue and plate
- Genomic prep of 477x475 - PCR with upp locus oligos
- Microscopy on J01022x408C cultures
- If you have time, run gel on digestions, purify, ligate and transform to make I716482 pbad.1106H.Cre
Samanthaliang 20:22, 2 October 2007 (EDT)
- Analyze sequencing
- Grow up colonies from the Cre transformations in Carb
- Grow up J01022x408C colonies with and without arabinose
- Collect Tecan data from upstairs - see if there's any visible phenotypes - BamHI and barnase hits!
- Grow up 462, 452, and 463 for a small scale competent cell prep (ask someone to seed them for you tomorrow!)
- Start digestions to make pbad.1106H.Cre
Samanthaliang 19:12, 1 October 2007 (EDT)
- Make a stock of arabinose-Carb LB for the Tecan assay as well
- Tecan growth ASSAY! - Start at 6pm!
- Grow up 2 colonies of 481
- Small scale competent cell prep of 475 and 408C
- transform in those plasmids 476&477 into I716475 - split the mix between an Amp plate and a Gen plate
- transform pSB3C6-J01022 into 408C - plate AmpCam
- Sent out 451-454 and 461-464 out for sequencing to confirm that they are right and not just mutants with misleading band sizes
Samanthaliang 16:46, 30 September 2007 (EDT)
- Grow up a 408C culture from the plate need to do a small scale competent cell prep for it
- Grow up a I716475 (lox.gen.lox KI) from the -80 - also need to do a small scale competent cell prep
- Grow up 96 block without arabinose for Tecan assay - don't forget controls DH10B
- Sign up for Tecan for Monday day or night
- Ligate and Transform 481
Samanthaliang 19:23, 28 September 2007 (EDT)
- Analyzed sequencing
- Resubcloning 481 because I got some strange sequencing results for it that made me check back to some old sequencing, and I realized that I commented on the sequencing wrong and actually used the wrong clone because my sequencing logs had that error. But everything is fixed now. I'll get the digestion and purification done today.
Ligate and transform on Sunday when I come in.
- Things are on hold a bit until I can come in 2 consecutive days (sunday&monday?)
Samanthaliang 17:53, 25 September 2007 (EDT)
- Look at Ara-toxin assay plates and document the results - unfortunatetly, there's pretty much no difference between the arabinose plates and the non-arabinose plates for plasmids I716451-454, I716461-464, but I think it's just a poor assay because the positive controls 408C and 408E didn't show any difference either. I think it's maybe because the growth stops a little bit later, or maybe there isn't enough arabinose in the plates. I think I have to do a Tecan assay.
- Show Jin and Mahdvi how to use the cytometer
- I won't be back in lab until Friday, so a lot of growing up stuff is going to have to wait.
Samanthaliang 20:04, 24 September 2007 (EDT)
- Analyze sequencing
- DO THE ARA-toxin ASSAY - take plasmids of the pbad-rbs-toxin type - dilute them about 20 fold (just to decrease the colony yield), heat shock them into DH10B's, then plate equal volumes of the material on amp or amp+ara plates. It should be readily apparent from the colony counts whether these guys are toxic or not.
- I'm also doing the same assay on 408C and 408E as positive control
- Pour Ara-Carb Plates
- Note - I'm missing a few entries in the wiki because I've been neglecting writing down what I'm doing, but basically I've been doing some subcloning in order to be able to knock a inducible toxin cassette into the genome that involves loxes and Cres.
- Note - I'm scrapping the whole building a lox.rbs.Cre.tt.lox because it's really really hard to do. I realized that even though I initially thought that all my subcloning woes had something to do with cloning methods because all of my constructs were just a single lox (which is one of the parent vectors) - after doing it several times with different methods (the last being dbbs) - I think I can come to the conclusion that there is probably enough background transcription of Cre that is causing recombination, which would also give me the product of Cre.
Samanthaliang 18:12, 15 September 2007 (EDT)
- Miniprep 445 and 446
- Analytic digest of 445 and 446 - sequence any that look good
- Water plants
Samanthaliang 18:12, 14 September 2007 (EDT)
- Grow up 445 and 446
Samanthaliang 18:04, 13 September 2007 (EDT)
- Subclone 445 and 446 using ddbs method
- Look for a repressible promoter
- check plates
Samanthaliang 20:26, 11 September 2007 (EDT)
- Spot the I716475xBca9096 and I716475xI716438 on Gen plates and Carb plates to select for colonies that have lost the GenR gene - totally overkill because not a library but whatever
- Analyze sequencing - bad again - starting ddbs method now - did the PCR
Samanthaliang 17:55, 10 September 2007 (EDT)
- grow I716475xBca9096 and I716475xI716438 in 96 well block to spot on plates
- send 445 and 446 things out for sequencing
- Look for more repressible promoters
Samanthaliang 18:14, 8 September 2007 (EDT)=
- take 200ul of each row and each column of the BglII methylase block and do a genomic prep on them. Digest with BglII, then run a gel
NOTE - I realized that I couldn't do what I bulleted above because the pzhu promoter doesn't work and in fact, just kills the cells. The pellets that Arthur were observing before were actually just salt precipitate. I can't believe he didn't catch that and I really wish that I had thought more about how weird his results were before I started subcloning with it.
Samanthaliang 18:14, 7 September 2007 (EDT)
- Analytic digest of 445 and 446s
- Set it out for sequencing
- Small scale competent cell prep of lox.gen KI I716475
- Transform I716438 and Bca9046 into the competent cells - plate on Amp
Note - The next day - grow them in a 96 well block, then the day after that, spot on Amp plate and Gen plate to see which have lost their function
- Grow up 36 colonies in a 96 well block of I716469 the BglII pzhu.rbs.methylase
Samanthaliang 19:49, 5 September 2007 (EDT)
- Right when you come in - grow up some I716475 to do a small scale competent cell prep so that 3 hours later, you can transform a plasmid with Cre into it and check to see if GenR is lost. (I716438 and Bca9046)
- Okay so the above point didn't work because it's been 3 hours and the cultures are still clear, which means that I probably won't be able to do the transformation tonight - Instead, streak the -80 stock of the knock in on a plate and ask Gabe to grow it up 3 hours before you come in tomorrow.
- Miniprep 24 pzhu.rbs.methylase cultures in zinc media.
- Digest zinc cultures with BamH or BglII and AlwnI (check to make sure the bands don't fall on top of each other) and see if there are 2 bands or 1 band on the gel. Select for 2 banders.
Note - Darn, the gels game out blank, which is really strange, but I guess maybe there is some toxicity to the zinc, because the cultures were pretty clear and not dense at all even though i left them overnight. I used no water in my digest, and all miniprep to compensate for the low concentration of the miniprep, but I guess it wasn't enough.
- Colony PCR on 445 and 446
Samanthaliang 19:14, 4 September 2007 (EDT)
- Grow 24 pzhu.rbs.methylase cultures in zinc media. (12cultures-470s, 6(1-6)cultures of 469-1, and 6(7-12) cultures of 469-2)
- Analyze sequencing - the 445 and 446s were wrong again
- Analytic digest of 445 and 446 clones that I did not sequence showed that they are probably not right as well
- Subclone 445 and 446 again using the BglI method instead of modified123 method - if this doesn't work, I'm not too sure what I'm going to do since it seems too big to do dbbs on.
Samanthaliang 15:09, 2 September 2007 (EDT)
- Digest I716469 and I716470 with BamHI/BglII, then retransform them to look for the ones that are protected (ask someone to take these plates out on Monday)
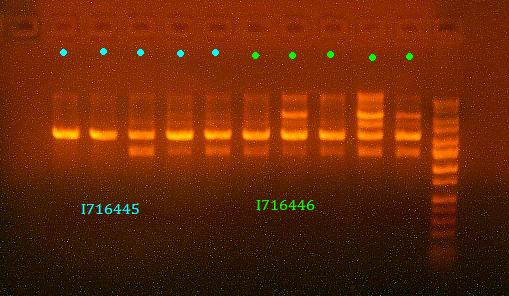
- Analytic digest of I716445 and I716446 clones - accidentally ran the gel too long, so can't verify for sure, will send out clones 2 and 3 of both for sequencing
- Send some I716445 and I716446 clones out for sequencing
- GENOMIC PREP of I716475 lox.gen.lox knock ins
- Expand PCR on lox.gen.lox knock ins with ca797F/R
- Run a gel on the PCRs
- All of the samples look correct (lox.gen.lox knock-in should be about 1kb and the upp locus would have been about 2kb)
- Make -80 stocks of lox.gen.lox I716475 clones that look good - all 3
Samanthaliang 15:58, 1 September 2007 (EDT)
- Didn't run a gel on the I716475 (lox.gen.lox knock-in) PCRs because I realized that I didn't do a genomic prep, but rather a miniprep so those PCRs would come back blank
- Miniprep I716469 and I716470
- Miniprep I716445 and 446 clones
- Grow up lox.gen.lox knock-ins again
Samanthaliang 18:12, 31 August 2007 (EDT)
- Grow up 445 and 446 colonies
- Chris grew up my knock-ins yesterday, so I miniprepped them today, start a PCR on them with ca797F/R to check check if the gene has been inserted
- Pool the pI716101-I716469 libraries (2), and the pI716101-I716470 library(1 didn't grow - the one using David's digest)
- Make LB agar
Samanthaliang 17:36, 29 August 2007 (EDT)
- Transfer 469 and 470 to a low copy plasmid
- Cloned 445 and 446 again - this time doing the ligase heat kill and extra digestion and the end
- Knocked in lox.gen.lox again - I forgot to add arabinose the first time
Samanthaliang 17:36, 22 August 2007 (EDT)
- Subclone 445 and 446 again now that 443-2 and 444-1 are confirmed
- Turn Vai'stuff into glycerol stocks on Wednesday
- Looks like the colonies are really tiny on the knockins plate - leaving them in the incubator a little longer to make sure that they are actually colonies and not just bubbles
Samanthaliang 14:20, 21 August 2007 (EDT)
- Analyze Sequencing - eek realized that 445 and 446 are not correct
- Knock-in lox.gen.lox into the Dh10B genome with pKD46
- Grew up pKD46 again just in case it doesn't work today
- Grow up colonies of 448, 459, and 460 - only 460 is good
- Pool 469 and 470 libraries (pzhuC.rbslib.methylase)
- Religate I716447 and transform - didn't get any colonies from yesterday - actually don't have to do this because 445 is not a good clone.
Samanthaliang 17:43, 20 August 2007 (EDT)
- Turn Vinni, Nhu, Hannah, and David's stuff into glycerol stocks
- Miniprep the 468 CamR and KanR pools
- Miniprep colonies of 466
- Miniprep colonies of 445 and 446
- Subclone pbad.rbs.atg.lox.rbs.Cre.TT.lox variants (447 and 448 with I716449, 459 and 460 with I716450) using 445 and 446 - as a note I used clones 2 of the green eppendorf variants
- Subclone pzuC.rbslibrary.methyl transferases (469 and 470)
- Analytic gel on 465 and 466 clones
- Grow up a pKD46 colony in 2YT with Carb(30 degrees!) - plate is in fridge now
- Send things out for sequencing
NOTE - After the pzhuC.rbs.methyl transferase variants are built and sequenced, need to move it to a low copy plasmid that David has (I716101). Then miniprep the entire library, cut with BglII or BamHI, then retransform it to select for ones that are protected. Then grow up 24 cultures in Zinc media, miniprep those, then cut with BglII or BamHI and another enzyme, and look to see if there is 1 band or 2 band as a negative screen.
Samanthaliang 18:23, 18 August 2007 (EDT)
- Digest the PCR product with DpnI, gel extract, and concentrate it
- Grow up colonies of 466
- Grow up colonies of 445 and 446
- Pool the 468 CamR and KanR plates
- Miniprep the 467 CamR and KanR pools
- Get someone to put stuff in the fridge for you until Monday
Samanthaliang 20:13, 17 August 2007 (EDT)
- Transform pKD46 (30 degree temperature sensitive) into DH10B - might need to electroporate but may be able to heat shock
- Pool 467 (CamR and KanR variants) clones - pick one that looks the best on the plate to proceed with the next step
- Miniprep Bca1149 clones
- Miniprep 465 clones
- Resubclone 466
- Subclone 445 and 446
- PCR the knockout piece
Samanthaliang 15:43, 16 August 2007 (EDT)
- Grow up 465 clones (466 positive control indicated that it was mostly parent vector)
- Miniprep the 443 and 444 clones
- Analyze sequencing for 438 and 439 - look good
- PCR basic part Bca1149 M.BamHI-2
Samanthaliang 15:43, 15 August 2007 (EDT)
- Analyze sequencing for 438 and 439 - Actually Quintara failed at getting sequencing back today so I'm just going to have to grow up a bunch of clones and figure out which ones i want tomorrow
- Grow up clones of 443 and 444
- Miniprep some of the 2 Bca9236 rbs library cultures - about 6 mLs
- Subclone 465, 466 - pzhuC.rbs variants (1106A, 1106C)
- Subclone Cam.rbs.BglII methyl transferase (467)
- Make biobrick Bca1149 (M.BamHI-2) so that I can builld I716468 which is Cam.rbs.BamHI
- Order oligos for knock in of lox.GenR.lox - get Chris to check them first
- Make slides for presentation - ah
Samanthaliang 18:47, 14 August 2007 (EDT)
- Miniprepped 438 and 439 clones
- Subcloned 443 and 444 (rbs.cre.tt.lox)
- Subclone 465, 466 - pzhuC.rbs variants (1106A, 1106C) - Nevermind - digest was not as expected and it's too late to start over now
- Transformed the Bca9236 rbs libraries into DH10B - straight into a 50ml culture
- Talked to Chris and John about direction - doing the 3 generation approach
Note - will have to eventually make pzhuC(Zinc repressible promoter).rbs.BamHI/BglII methyl transferase and move it to a low copy plasmid - will put it in equilibrium with BamHI and BglII
Note - may have to start doing knock-ins with the lox sites
Samanthaliang 18:06, 13 August 2007 (EDT)
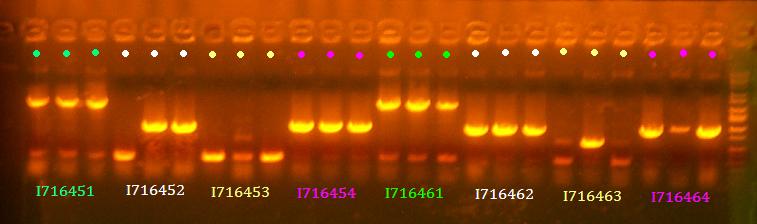
- Miniprep 451-1, 452-2, 453-1, 454-1, 461-1, 462-1, 463-2, 464-1
- Grow up 438 and 439 clones
- Plan a Tecan experiment for troubleshooting the toxins - putting this on hold until I figure out everything else I'm doing first
Samanthaliang 18:06, 12 August 2007 (EDT)
- Sublconed 438 and 439 using John's modified 123 method
- Grew up 451-1, 452-2, 453-1, 454-1, 461-1, 462-1, 463-2, 464-1
Samanthaliang 21:10, 11 August 2007 (EDT)
- Started to make 438 and 439 but while running the gel on a digest, the gel was too thin and my samples fell through and were lost and there wasn't enough to do another digest.
- In future, check the gels to make sure they are thick enough or else pour your own stock
Samanthaliang 17:07, 10 August 2007 (EDT)
- Miniprep lefty and righty things
- Ran gel of Colony PCRs and took plate out of the incubator
Samanthaliang 17:07, 9 August 2007 (EDT)
- Grow up lefty and righty things
- Colony PCR of 451-454, 461-464 clones
Samanthaliang 17:07, 8 August 2007 (EDT)
- Miniprep Bca1114 and return one to Chris' stock box
- Miniprep 445 and 446 clones
- Analytic digest of 445 and 446 clones - bad, I just checked the sequencing for everything and it turns out that rbs.Cre.TT was never properly made, so clones 443 and 444 are actually rbs.Cre.lox clones which is not what I want, therefore I have to go back a few steps and try different cloning methods
- Subclone pbad.1090.atg.toxin variants (451-454) and pbad.1106A.atg.toxin variants (461-464)
- Transform 436, 437, Bca1114, 449 and 450 into Lefty for another approach at making rbs.Cre.TT
- Transform Bca1092 and Bca1114 into Righty
Samanthaliang 14:36, 7 August 2007 (EDT)
- Send out lots of things for sequencing (don't forget to use the reverse oligo or the ca56 oligo for several of them)
- Grow up clones of Bca1114
- Grow up 445 and 446 clones
- Maybe grow up 455 through 458 things in 96 wells to do a tecan assay with
- Definitely create an excel file with all the biobricks that you've made
Samanthaliang 21:58, 6 August 2007 (EDT)
- Retransform lox biobrick Bca1114 because it is running out
- Analytic digest of I716443 and I716444 showed that clones 2,3,4 looked good for 443 and clones 2 and 4 looked good for 444
- Subclone I716445 and I716446 from clone 2s of the parents
- Miniprepped all clones of 443, 444, 450, 449, 455, 456, 457, and 458
- Analytic digest of all
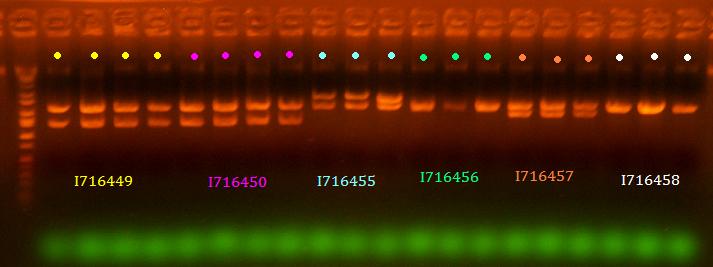
lane 1 is ladder, lanes 2-5 are 449, lanes 6-9 are 450, lanes 10-12 are 455, lanes 13-15 are 456, lanes 16-18 are 457, lanes 19-21 are 458 (All digested EcoRI/BamHI)
Samanthaliang 21:58, 5 August 2007 (EDT)
- Grow up clones of I716450 and I716449, and I716455 through I716458
- Grow up clones of I716443 and I716444 (rbs.cre.tt.lox variants)
Samanthaliang 16:03, 3 August 2007 (EDT)
- Miniprep the I716438 and I716439 clones
- Analytic digest of the I716438 and I716439 clones
- Send out for sequencing
- Subcloning rbs.Cre.TT.lox if any of the 438 and 439's look good
- Colony PCR I716450 and I716459, and I716455 through I716458
- The colony PCR came out blank again strangely enough but I don't think this means my samples are necessarily wrong. I might just have to grow a bunch of stuff up on Sunday and do analytic digests instead.
- Analytic digest of the I716442 cassettes to check for recombination
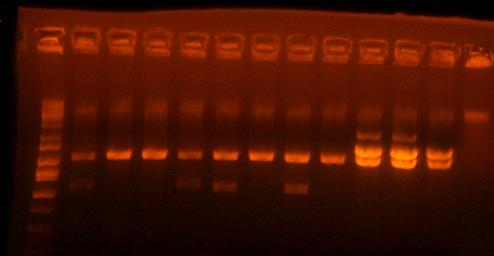
the first lane is the ladder, the next four are I716438 clones, the next four are I716439 clones, and the next 3 are I716442 clones. Although the top band does not correlate perfectly for any of the I716438 and I716439 clones, I think they are worth sequencing and going onto the next step of subcloning just in case. I've saved clones 1 and 4 of 438 and clones 1 and 3 of 439. Also clone 3 of 442 looks good, but i'm sending both clone 2 and 3 for sequencing.
- Get someone to take plates out of incubator and put into fridge on saturday - will come in sunday to grow up stuff
Samanthaliang 18:42, 2 August 2007 (EDT)
- Build pbad.1106A.ATG - I716450
- Build pbad.1090.ATG - I716459
- Build pbad.1106A.toxin(no ATG) - I716455-I716458
- Note - I716457 the barnase variant already exists as part I716408C or E
- Miniprep Bca1092s (TT) for future use
- Miniprep I716442 variants
- Grow up 438 and 439 cultures (4 clones of each)
Samanthaliang 18:42, 2 August 2007 (EDT)
- Analyze sequencing
- Colony PCR of 442 clones and 439 clones
- Grow up colonies of 442 with arabinose (PCR tomorrow to see if any crazy recombination has happened)
- Grow up clones of Bca1092
- (438 didn't have any colonies) and the 439 clones failed the Colony PCR that I did
- Subclone 438 and 439 again - redigest everything - this time the terminator is right for sure, so now the other digests should be good too
Note - So I talked to Chris and asked why something like pbad.rbs.toxin wouldn't work and it's probably because the phenotype we want is too fragile and difficult to obtain in this manner and that working in the area of Cre, where people really want to figure out to make that work, is a good area to work in so hopefully I can get that cassette to be stable and work as an effective switch. However if there isn't enough time before igem, something easier could work, so right now I'm going to try to make just pbad.rbs.toxin and pbad.rbs.ATG.toxin variants to see if i can get anything that has the phenotype we want. Also it isn't a bad idea to kill the cells and look at them under a microscope to see what they look like when they're dead. If the cells lyse then we don't want it, but it'd be interesting to see how the restriction enzymes differ from the general nucleases and from the ribonucleases. I wonder if they could be potentially lysing, especially if their cell wall breaks down and they are unable to regenerate since their dna is bad.
Samanthaliang 17:51, 31 July 2007 (EDT)
- Colony PCR of 438 and 439 - I think one of the oligos I used is not quite right, maybe too dilute or something because all of my colony PCRs came back blank and I know this happened to Austin and Arthur too before and the only thing I did differently this time is that I used a different aliquot of oligo.
- Miniprep everything you grew up yesterday
- Subclone 442 out of 440 and 441
- Try redigesting Bca1092 (TT) from a different aliquot since the one I've been using yields 4 bands instead of 2 bands, although previously I've been cutting out the band that is the right size - It worked
- Subclone 438 and 439 again
- Retransform Austin's small aliquot of Bca1092
- Send out for sequencing
==Samanthaliang 19:11, 30 July 2007 (EDT)==
- Colony PCR of 438 and 439 (5 clones of each)
- Grow up ones that pass the test (none really do so I'm resubcloning, but also growing up a few)
- Colony PCR some clones if I716441 and I716440
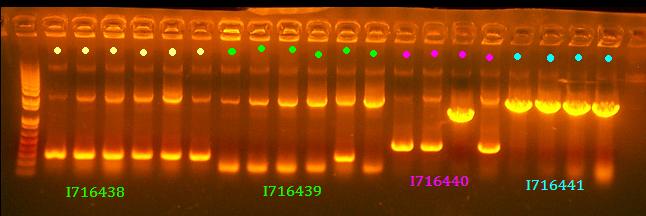
No 438 and 439 clones look right and I think it's rather strange that there are 2 bands for most of them although it is possible to have some recombination with Cre. All clones of 441 look good and clone 3 of 440 looks good.
- Resubcloning 438 and 439.
- Pool I716430 through I716435 so you have minipreps of them
Samanthaliang 14:30, 27 July 2007 (EDT)
- Evaluate sequencing - clone 4 of 436 and clone 1 of 437 are good
- Subclone rbs.Cre.TT with 436 and 437 to make I716438 and I716439
- Subclone Cre.(barnase cassette) with 212 and 240 to make I716440
- Subclone pBad.1106A with Bca1109 and Bca1106A to make I716441
Samanthaliang 19:05, 26 July 2007 (EDT)
- Colony PCR on the cultures of 408A, 408D, 411A, 434A, and 412A with and without arabinose (maybe will see some recombination and shorter PCR products for ones with arabinose that show some phenotype)
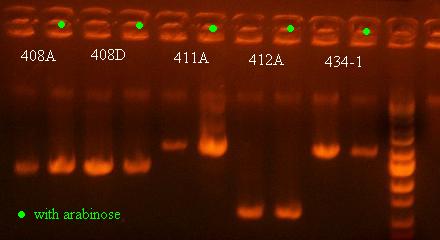
There was no change in the size of the PCR product for any of these, which I actually started to expect because the cells that are most prominent in the saturated culture will not be the cells that have undergone recombination and died already. Although it is interesting how low the bands are for 412A, which means that some recombination must have occurred for that variant as well (T.1.BglII), showing that other structures can show a recombination phenotype as well.
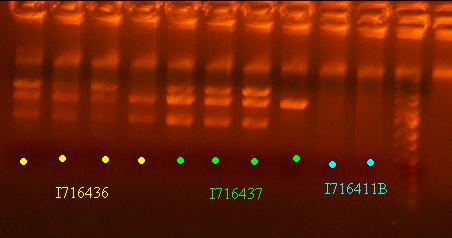
- Miniprep the 436 and 437 clones and do a quick analytic gel
- Send out 436 and 437 clones out for sequencing that pass the analytic gel test - clones 2 and 4 of I716436 and clones 1 and 2 of I716437
Samanthaliang 15:15, 25 July 2007 (EDT)
- Make large scale competent cells of Righty and Lefty
- Pick colonies if I716436 (1106A.Cre) and I716437 (1109.Cre) - 4 of each
- Grow cultures of 408A, 408D, 411A, 434A, and 412A with and without arabinose to see if there is any recombination induced by arabinose of clones that have full cassettes without being induced and yet show a slight decrease in growth from the tecan assays (under 0.95 OD at the end)
- Grow up cultures of 411B since it grew on the colony PCR plate unexpectedly. It could just be contamination or an escaped mutant but hopefully it's what I want. Also, it showed no bands on the colony PCR which is weird since it ended up growing on the plate
NOTE The new thing that i want to build is:
.pbad.rbs.atg.lox.rbs(1106A).Cre(GTG).TT.lox.barnase.
This way we can show that rbs.cre.tt is stable, then show lox.rbs.Cre.TT.lox is stable, and then focus solely on the switch by measuring the size of the cassettes and eliminate complicating studying the switch by adding toxins into the mix. After making sure the switch works by adding pbad and colony PCRing on a few clones, then add the toxin into the mix and that way we will be able to isolate the problem.
Samanthaliang 19:09, 24 July 2007 (EDT)
- Subcloned 1106A.Cre and 1090.Cre (rbs.Cre)
- PCR with ca56(in pBad promoter)/G01001 on minipreps of 416, 417, 418, 401, 411, and 412 to examine the size of the cassette - these are the ceaB variants and also a barnase and 2 bglII variants that showed some activity in the initial round of tecan screening
- Colony PCR of inactive (or rather less active) 408s - 408A, 408B, 408D, 408F
- The wells that I grew yesterday with the RFP reporter mechanism did not turn red at all so I'm going to examine that some more.
- Colony PCR of of 434 (GTG TT verision of RFP reporter) - 4 clones to look at cassette
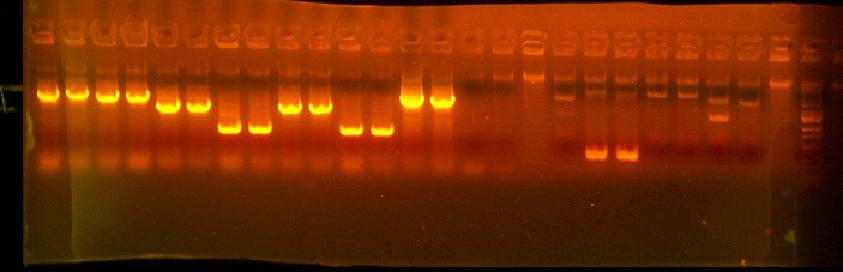
This is annotated in my paper notebook.
- Chris saw that rbs.atg.bamH and rbs.atg.BglII wasn't lethal in Lefty, Righty, and DH10B - but will be checking if it is lethal once he adds a promoter
- Grew up cultures of Lefty and Righty
Samanthaliang 21:28, 23 July 2007 (EDT)
- Pick colonies from I716430 through I716435 in a 96 well block (swirl each colony in a well with arabinose and a well without) then check if the cultures turn red (might even be able to run a quick tecan on it to see the RFP levels)
- Analyzed sequencing results for 408C and 408E - unfortunately, these two guys are the same thing - [pbad][rbs][barnase - no ATG], which is not what I want because it is missing both Cre and the cassette. This must have occurred by some crazy recombination or something. I need to do some PCR mapping on some other clones to troubleshoot. I think hits are just falling out like crazy. Maybe it'd also be helpful to make libraries on low copy plasmids? Might be able to pick up more hits.
- Long google chat with Chris about next steps
Samanthaliang 15:17, 20 July 2007 (EDT)
Things to read about:
- Does barnase have DNase activity? Haven't really found anything saying that it has DNase activity yet, but it is a ribonuclease that destroys all the RNA and targets the ribosome, which in turn, stops DNA synthesis such that those bacteria should no longer be able to replicate and grow.
- Is there a way to do dapi (pronounced dap-E) measurements for e.coli? dapI binds to DNA, wonder if can use a cytometer or something? Do a dapi stain? YES! Can do a dapi stain in e.coli and then do measurements with a flow cytometer.
- Make sure that we're not just making a toxic protein or that Cre itself is toxic or something like that.
So I'm having a lot of trouble isolating the plasmid of 411B. The plate that I got from streaking a 96 well culture has really sickly single colonies that are super tiny. I can't get any of the colonies that I pick to grow in liquid media even when I add glucose. It also didn't grow after being streaked onto another plate. It seems like that in order to try to isolate a plasmid and sequence it, I'm going to have to take a swab of the plate in a dense area and miniprep that (put it in p1 buffer directly) then I'm going to have to sequence off of a PCR product.
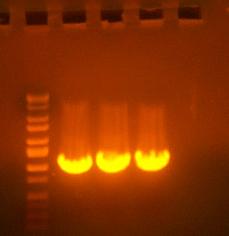
Here's a picture of the gel:
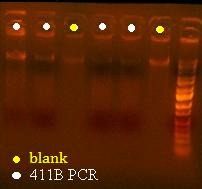
Unfortunately, there were no bands on it, meaning that either that part isn't what I think it is, or else, all the cells are already dead and the primers weren't able to pick up anything big enough to see on the gel from the plasmid. I can also see kind of a smear, which could mean that I'm picking up junk, but more likely, it's just some discoloration from the dye or something.
- Miniprep 408C, 408E, and 411B (by swabbing it)
- run PCR on 411B (ca998/G01001) and gel purify it
- Sequence 408C, 408E
- Make -80s of 408C and 408E
- Look at spotted plates - the spots actually look like they're less dense for the ones that showed a plateauing growth rate. I don't think it's obvious enough to be convincing of anything though. I guess if any cell is alive though, it would grow in the spot.