Turkey/ Possible Projects
From 2007.igem.org
Project 1 Mexican Wave
FIRST VIEW
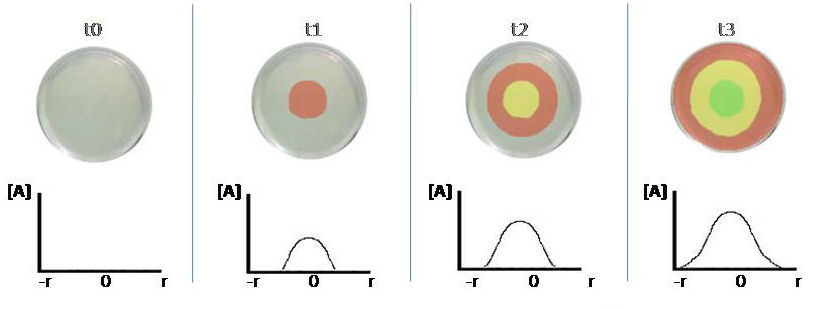
- Our first thought was to create a pattern by the change of colors in a concentration dependent manner; so that the production of a protein at the end of a different event would trigger the pattern formation, just like a goal triggering a *Mexican wave in soccer games.
- The plan was to have a plate having a lawn of same type E. coli cells having 3 constructs. Each construct would have a promoter activated by a different level of the initial signal protein and a reporter fluorescent protein coding part.
- The triggering protein A is being dropped on the middle of the plate, creating a concentration gradient on the plate as in the figure. Acording to this plan the color change will be once, and then by the rising level of [A], the plate will all become green.
SECOND VIEW
- The second view was to make this color change continuous and independent of a created concentration gradient. So first idea came up to be coupling this oscillation to an already oscillating protein, maybe to cell cycle. But then this oscillation will be observable only on a single cell unless we synchronize the cell cycles of different cells on the plate.
Project 2 Chase Simulation
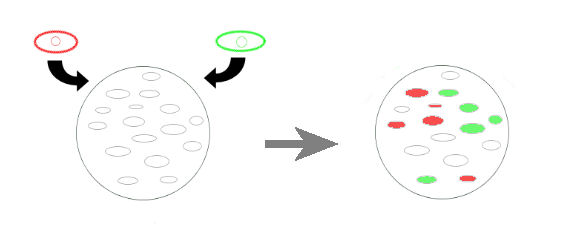
- The first idea of project was to simulate the competition between two different warrior cell types to invade a plate having a lawn of passive cells. The green and red cells are the invaders, and they represent two teams. The 'empty cells' are the ones to be invaded. Empty cells have two constructs, they are normally off and each construct can be activated by a red or green cells, invaders. If one of the constructs in empty cells is activated, this construct irreversibly closes the other constructs.
- The trick point in this project is to have two promoters which are normally off, and also repressed and activated by distinct proteins. There should be no leakage, because a little leakege in one of the constructs closes the other irreversibly. So we created a less complex version;
- We reduced the invaders to one type, and each team will have a separate plate. This invader will activate a normally off promoter which codes for a flourescent protein. In the registry there are there pairs of promoter-activator for this purpose. But the experience info is inadequate, so we will try all of them. This way we can overcome the leakage problem, if there is a constitutive leakege, there will be stable expression of the protein unless the invader is on action. So we can make a base level correction to see the effective contrubution of invasion.
- The Three pairs of promoter-activator are:
Part Info Plate Well Plasmid Length Cell Sel. BBa_B0015 Double terminator 1 1I pSB1AK3 3189 3318 445 V1001 AK BBa_B0015 Double terminator 3 3O pSB1AK3 3189 3318 445 V1010 AK BBa_b0034 RBS 1 3O pSB1A2 2079 2091 250 V1004 A BBa_I14032 constitutive promoter 2 17K pSB2K3 4425 4462 353 V1006 K BBa_E0020 E-CFP 1 7C pSB1A2 2079 2802 961 V1001 A BBa_E0030 E-YFP 1 7G pSB1AK3 3189 3912 1039 V1003 AK BBa_E0044 GFP 1 5B pSB1A3 2157 2916 1075 V1001 A BBa_J06504 mRFP 2 7N pSB1A2 2079 2793 952 V1009 A BBa_I12007 lambda Prm promoter activated by cI 2 15K pSB2K3 4425 4507 398 V1006 K BBa_C0051 cI repressor from E. coli phage lambda (+LVA) 1 5G pSB1A2 2079 2829 988 V1004 A BBa_R0062 promoter activated by HSL&LuxR 1 9G pSB1A2 2079 2134 293 V1004 A BBa_C0062 luxR repressor/activator 1 7A pSB1A2 2079 2835 994 V1004 A BBa_C0061 Synthesizes 3OC6HSL, which binds to LuxR 1 5O pSB1A2 2079 2697 856 V1004 A BBa_R0079 Promoter (LasR & PAI regulated) 1 19N pSB1A2 2079 2236 395 V1001 A BBa_C0179 LasR activator 1 3J pSB1A2 2079 2802 961 V1003 A BBa_C0078 autoinducer synthetase for PAI 2 7I pSB2K3 4425 5067 958 V1004 K BBa_R0080 Promoter (AraC regulated) 1 21L pSB1A2 2079 2228 387 V1001 A BBa_C0080 araC arabinose operon regulatory protein 2 7M pSB2K3 4425 5340 1231 V1004 K BBa_I1051 Lux cassette right promoter 1 18P pSB1A2 2079 2147 306 - A