USTC/Repressor Evolution on Plates
From 2007.igem.org
m (→Repressor design) |
m (→Repressor design) |
||
| Line 36: | Line 36: | ||
== Repressor design == | == Repressor design == | ||
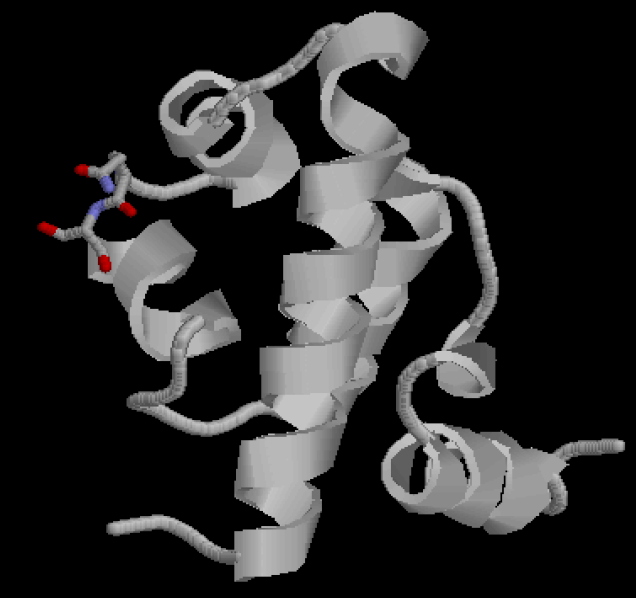
| - | [[Image:USTC_lac_mutation_sites.png|thumb | + | [[Image:USTC_lac_mutation_sites.png|thumb|'''Figure 2''' The recognition region of Lac repressor of which the amino acid residues we will try to modify.]] |
Figure 2 shows the recognition region of which the amino acid residues we will try to modify. We attempt to minimize the binding energy of DNA and protein between the recognition region and the target operator to find the optimal composition and arrangement of the amino acid residues of the recognition regions. | Figure 2 shows the recognition region of which the amino acid residues we will try to modify. We attempt to minimize the binding energy of DNA and protein between the recognition region and the target operator to find the optimal composition and arrangement of the amino acid residues of the recognition regions. | ||
With this computational method in silicon, we are able to screen thousands of proteins in a faster and cheaper way, compared with the traditional wet experimental methods. | With this computational method in silicon, we are able to screen thousands of proteins in a faster and cheaper way, compared with the traditional wet experimental methods. | ||
Revision as of 06:19, 2 August 2007
Contents |
Why to design repressor-operator pairs
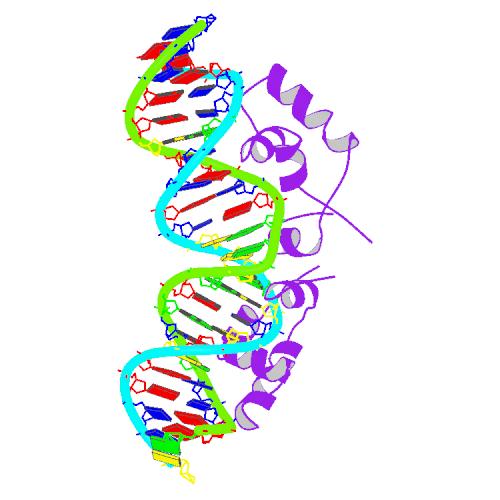
We have been attempting to construct several artificial repressor-operator pairs to serve as the connecting wires of our system, based on the knowledge of LacR and its binding site, and by means of protein design. We decide that they should be newly designed for three reasons,
- The number of natural regulators well studied is limited.
- Natural regulators do have some disadvantages.
- For example, it is well known that there are dozens of downstream regulatory sites of CRP in E.Coli, and if we abuse the CRP, some other natural pathways in the host bacterial will probably be interrupted.
- There may be several unknown sites to be bound with the selected native activators, and we might get unexpected results in such situations.
Start: Lac repressor
[http://en.wikipedia.org/wiki/Lac_repressor From Wikipedia]:
The lac repressor is a DNA-binding protein which inhibits the expression of genes coding for proteins involved in the metabolism of lactose in bacteria. It is active in the absence of lactose, ensuring that the bacterium only invests energy in the production of machinery necessary for the uptake and metabolism of lactose when lactose is present. When lactose becomes available, it is converted into allolactose, which inhibits the lac repressor's DNA binding ability.
The lac repressor protein has three distinct regions:
- a core region (which binds allolactose)
- a tetramerization region (which joins four monomers in an alpha-helix bundle)
- a DNA-binding region (in which two LacI proteins bind a single operator site)
The lac repressor occurs as a tetramer (four identical subunits bound together). This can be viewed as two dimers, with each dimer being able to bind to a single lac operator. The two subunits each bind to a slightly separated (major groove) region of the operator. The promoter is slightly covered by the lac repressor so RNAP cannot bind to and transcribe the operon.
The DNA binding region consists of a helix-turn-helix structural motif.
Selection of operator sequence
By means of bioinformatics we can select a DNA sequence that has never appeared in the genome of E.Coli, and let the regulator bind to the sequence with quite high specificity,. Therefore, we will not have to worry about the regulator’s interrupting the normal functioning of the host genome.
O1 aattgtgagcgctcacaatt O2 aattgtaagcgcttacaatt O3 aattgtaaacgtttacaatt O4 aattgtgaacgttcacaatt
Repressor design
Figure 2 shows the recognition region of which the amino acid residues we will try to modify. We attempt to minimize the binding energy of DNA and protein between the recognition region and the target operator to find the optimal composition and arrangement of the amino acid residues of the recognition regions.
With this computational method in silicon, we are able to screen thousands of proteins in a faster and cheaper way, compared with the traditional wet experimental methods.