Vaibhavi Umesh Notebook
From 2007.igem.org
My Construction Files
My Sequencing Files
~~!~~
notes
to do
Vaibhaviu 19:02, 1 June 2007 (EDT)
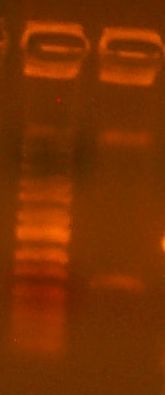
Today we cloned an RFP basic part into pBca9145. So, we did the construction file for pBca9145-Bca1145. We did a PCR of the RFP gene according to the construction file, and the gel is here:
It's a little dilute, but it should work nonetheless. We also digested the vector according to the construction file:
We gel purified the larger of the two bands. It looks like the digest went pretty well, as we see no single-cut plasmid DNA. We took it all the way through to transformation into DH10B cells today.
Vaibhaviu 18:45, 5 June 2007 (EDT)
Today we first observed the transformed plates that grew over the weekend. Four large, white, colonies surrounded by several satellite colonies were observed. No red colonies were observed which meant no parent vector had bled through onto the plate. FOr each colony, we made overnight cultures and placed them in the 'Incushaker'. Chris had already simulated this and two samples of the cultures were already ready for miniprep. We simulated a mini prep experiment and isolated the plasmids from the two culture samples. We then performed the analytical test digest and used BglII and XhoI restriction enzymes. These enzymes were chosen because they gave a distinct digestion pattern when compared with the parent vector.
Then, we simulated a colony PCR on one of the colonies on the plate, chosen randomly. We ran the results of both the digest and the PCR on a gel. The gel contained 5 wells. The first one contained a marker (the ladder), the second one The results of the gel were not very clear. THe parent vector and the PCR lanes both did not show up/ did not display the expected results. THe pictures taken were unclear, but when observed on the UV plate, the bands were brighter, but no additional bands showed up.
We also made the samples to be sequenced. We placed the mini prep plasmids in a 1.5 mL epp tube. We filled out the spreadsheet to send to Quintara and placed the samples in the metal box outside to get picked up. The specific oligos to be used were also mentioned in the spreadsheet.
Vaibhaviu 18:45, 5 June 2007 (EDT)
Today, we got the sequencing results from Quintara
Vaibhaviu 19:02, 12 June 2007 (EDT)
Today I got my sequencing results back. SodC #1 results were not very good and quintara was going to re-send them after sequencing them with a stronger signal. SodC #2 was fully correct. However, at base pair # 549 (on the sequencing results), there was a mis-read of 3 C’s when only two peaks could be seen on the graph. For methAP gene results, methAP#1 yielded a better result than methAP#2. However, although all the base pairs aligned for methAP#1, there was one situation at base pair # 762 (on the sequencing results), where the sequencing result had inserted an extra G not present in the original gene. When the graph was observed, the resulting peak was quite ambiguous and it was difficult to distinguish whether the peak was actually two separate peaks, or was three. As a result, just to double check, the plasmid was sent in for reverse sequencing using the reverse primer G00101 (the reverse primer of ca998). The results for this sequencing file will help establish that the mis-match and presence of an extra G was due to the ambiguity of the graph rather than the presence of an actual mutation.
VU005.MetAP#1 is the sample I sent in along with the primer G00101
Vaibhaviu 19:02, 13 June 2007 (EDT)
Since I had a sample of the SodC gene (#2) which was had perfect sequencing results…I saved that clone on the clonesaver sheet.
Apart from that, today was a really long day because I proceeded to attach ribosome binding sites to the SodC and Map genes I had sequenced. Most of the morning was spent in planning out the procedure as I had to identify the restriction enzymes to use to cut the gene and rbs and attach them together. This was the result of the planning:
RBS: pBca 9145 – Bca1090 SodC gene:
- cut with AlwNI ‡ 1519
- cut with BglII ‡ 1081
9145-Bca1090
- cut with AlwNI ‡ 1538
- cut with BamHI ‡ 553
2nd insertion: RBS: pBca1101 – Bca1128 SodC gene:
- cut with BglI ‡ 923
- cut with BglII ‡ 1677
Bca1128:
- cut with BglI ‡ 1010
- cut with BamHI ‡ 1990
MetAP (map) gene + 1128
MetAP gene:
- cut with BglII ‡ 1950
- cut with BglI ‡ 923
1128
- cut with BamHI ‡ 1990
- cut with BglI ‡ 1010
However, the 1090 rbs miniprep barely had any left in the tube, so I had to transform what was left of the miniprep and grow it out to make some more to use. So I transformed a plate with the 1090 rbs. However,
Vaibhaviu 19:02, 14 June 2007 (EDT)
Today I observed the three plates I had transformed yesterday. There were many colonies on the plate, especially the ones with the ligations on them, making it difficult to isolate individual colonies from the plate. However, I picked three colonies from the plate with the ligations and one colony from the 1090 plate. However, I cannot grow them until 5:00 today because I will not be able to come in before 9 the next day and it is not advisable to exceed 16 hours for growing the colonies. However, as prepartion, the tubes were labeled and the media was poured into them. Now, I examined both the reverse sequencing results for the MetAP #1 gene and the SodC #1 gene which came in. Both the sequences were perfect, without a single error. I saved both these clones on the clonesaver incase I need to use them to make a mini-prep in the future. Since the reverse sequencing results for the MetAP #1 gene were perfect, it can be concluded that the one mismatch error present in the forward sequencing file was a sequencing error rather than an error in the clone.
Since the MetAP #1 gene sequencing results were perfect, I will proceed to attach an rbs to the gene. In order to do this, I need to plan the construction file for the MetAP #1 gene and the 1128- 051/052 rbs plasmids. However, it was found the best sites were those that were cut with BglI and BglII/BamHI, which was the same combination used to digest the rbs site yesterday. As a result, I already had a sample of the rbs site available that had been digested and was ready to use in ligation. So, I went ahead and digested the MetAP #1 gene, ran a gel, extracted the 2kb band and purified, I eluded with 10microliters of water. Then, I ligated the insert along with the MetAP #1 vector bands. After ligation, I transformed the mixture.
One problem encountered was that during the gel purifications step, I made an error and did not label the zymo column I was using to purify the the MetAP #1 gene. I had used another empty zymo column with water to balance it in the centrifuge. Thus, after spinning it once, I no longer knew which zymo column had water, and which held the DNA. Thus, I was forced to continue purification with both zymo columns and ligated the sample from each column with either the 051 insert or 052 insert as I did not have enough of the insert to ligate both samples with both inserts. I also transformed both ligations onto a plate. If all goes well, one of my transformations will NOT yield any colonies and that will be the sample that had contained the water. The other transformation should yield colonies which shows that the bacteria had successfully taken up the plasmid formed during ligation (containing the composite part of the gene+ rbs). The results seen tomorrow will determine the identity of each ligation solution.
Vaibhaviu 19:02, 15 June 2007 (EDT)
I observed the plates I had transformed yesterday. Unfortunately, the plates – one with MetAP #1 and water, and the other with MetAP #1 and and the rbs site both grew colonies which meant my control (using water in the ligation) had failed. I tried trouble shooting by figuring out what went wrong. One possible explanation was that the plasmids had only been singly cut and binded back together during ligation. However, instead of doing a colony PCR to figure out what went wrong, it seemed easier to redo the process. Also, I had to re-do the entire rbs attaching process because I had found out I had used the wrong sequence file to plan the process meaning the restriction enzymes I had used would not work with what I actually had.
I understood the difference between using single rbs’s and rbs libraries. The rbs library had a promoter, and rbs site, and an RFP in the plasmid. However, specific libraries had the promoter replaced with different antibiotic markers. The 51 library had a kanamycin antibiotic marker, while the 52 library had the Cam marker. I also found out that the genes Arthur and I had been working on needed to be paired as they performed a similar function. I was planning on attaching a single rbs and an rbs library to each gene that I was using.
Single strong rbs sites I could use:
- 1090
- 1106A
- 1106B
- 1128A
The rbs libraries I could use:
- pBca1101 – I716051 (kanamycin resistance)
- pBca1101 – I716052 (cam resistance)
The genes would be paired in the following way:
- KatG + SodC ‡ hydrogen peroxide
- Map + AHSP ‡ hemoglobin
AHSP was the synthesized gene that austin had cloned into Bca9145. Today I focused on Map + AHSP and planned out how I would attach rbs sites to these genes.
Map gene + 1090
| map gene | 1090(rbs) |
|---|---|
| BglII - 1950bp | BglI - 942 |
| BglI - 923 | BamHI - 9 |
| XhoI - 1140 (this fragment will not ligate) |
| map gene | 1090 (rbs) |
| BglII – 1950|BglI – 942| | |
| BglI – 923|BamHI – 9| | |
|
Map gene + library (51) Map gene: 51 (rbs library) BglII – 2864 BamHI - 2904 EcoRI – 9 EcoRI - 1065 AHSP + 1090 AHSP gene: 1090 (rbs) AlwNI – 1519 AlwNI - 1538 BglII - 868 BamHI - 553 AHSP + library (52) AHSP gene: 52 (rbs library) BglII – 2378 BamHI - 2904 EcoRI – 9 EcoRI - 933 I had to leave a little early due to a dance rehearsal that had been scheduled so this was as far as I got today.
Vaibhaviu 19:02, 18 June 2007 (EDT)Today I started the process of attaching rbs sites to the Map + AHSP genes I had planned out on Friday. I followed the plan and digested the genes, ran a gel and extracted the bands. I gel purified and put the sample in the freezer. I would finish the process tomorrow.
Vaibhaviu 19:02, 19 June 2007 (EDT)Today I continued to ligate and transform the stuff I had started yesterday. After I finished transforming the plates, Arthur gave me the katG gene in the 9415 plasmid so I will first plan out the digestion for both the katG gene and the sodC gene, perform a restiction digest, run a gel, and extract the bands and gel purify today. That way, I can come in tomorrow and ligate and transform. both the katG gene and the sodC gene.
1090: Digest with AlwNI: 1538 Digest with BamHI: 553 SodC + library (51) SodC: Digest with EcoRI:9 Digest with BglII: 2591 51 (library): Digest with EcoRI: 1065 Digest with BamHI: 2904 KatG + 1090 ‡ 1-2-3 method KatG: Digest with: Digest with BglII: 1090: Digest with: Digest with BamHI: KatG + library (52) KatG: Digest with EcoRI: 9 Digest with BglII: 4250 52 (library): Digest with EcoRI: 933 Digest with BamHI: 2904
Vaibhaviu 19:02, 20 June 2007 (EDT)Today I started the first step of the 1-2-3 method to attach the 1090 rbs to the katG gene. I had to transform the katG gene miniprep into the righty strain and the rbs (1090) miniprep into the lefty strain. I plated both of these. I also digested and gel purified the following SodC & 1090 KatG & 52 SodC & 51
Vaibhaviu 19:02, 21 June 2007 (EDT)Today I continued what I was doing yesterday…tying up a lot of loose strings! I had transformed the lefty and righty plates yesterday. I got colonies, however, they were not very uniformly spread out..which might have been a result of the way I had spread the culture on the plate. I was able to isolate two colonies on each plate and I grew them in the incushaker. Then, the plasmids I had gel purified yesterday needed to be ligated. I ligated the fragments and proceeded to transform them.
Vaibhaviu 19:02, 22 June 2007 (EDT)LAB FLOODED…no work done!
Vaibhaviu 19:02, 25 June 2007 (EDT)
Vaibhaviu 19:02, 26 June 2007 (EDT)
What I sent in for sequencing: VU006 – map + 1090 #1 VU007 – map + 1090 #2 VU008 – AHSP + 1090 #1 VU009 – AHSP + 1090 #2 VU010 – SodC + 1090 #1 VU011 – SodC + 1090 #2
Tomorrow’s plan:
Vaibhaviu 19:02, 27 June 2007 (EDT)
Gene + rbs Where it is What needs to be done immediately Work for the future Map + 1090 Miniprepped (3 samples) Sequencing Transformed into lefty strain Map + 51 AHSP + 1090 Miniprepped (3 samples Sequencing Transformed into righty strain AHSP + 52 SodC + 1090 Miniprepped (2 samples) Sequence needs to be analyzed Needs to be transformed into lefty SodC + 51 Miniprepped ( 1 sample…CORRECT) KatG + 1090 Miniprepped (2 samples) Sequencing Transformed into righty strain KatG + 52 Miniprepped (1 sample…CORRECT)
VU012 – katG + 1090 #1 VU013 – katG + 1090 #2 VU014 – map + 1090 #1 VU015 – map + 1090 #2 VU016 – AHSP + 1090 #1 VU017 – AHSp + 1090 #2
LEFTY RIGHTY Map + 1090 AHSP + 1090 SodC + 1090 KatG + 1090
|