|
|
| (242 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | [[Image:Eth_zh_logo_2.png|800px]] __TOC__ | + | [[Image:ETHZ_banner.png|830px]] |
| - | == Team Members ==
| + | <!-- |
| - | <div style="float:right"> | + | <center>[[ETHZ | Main Page]] [[ETHZ/Model | System Modeling]] [[ETHZ/Simulation | Simulations]] [[ETHZ/Biology | System Implementation]] [[ETHZ/Biology/Lab| Lab Notes]] [[ETHZ/Meet_the_team | Meet the Team]] [[ETHZ/Internal | Team Notes]] [[ETHZ/Pictures | Pictures!]]</center><br> |
| - | {|
| + | --> |
| - | |- | + | __NOTOC__ |
| - | | class="taxo-image" | [[Image:|thumb|398px|'''Picture:''' The ETH Zuerich iGEM Team 2007 ''(left to right: )'']] | + | <html> |
| - | |}</div> | + | <script type="text/javascript" src="http://christos.bergeles.net/eth_dropdowntabs.js"> |
| | | | |
| - | For the International Competition on Genetically Engineered Machines, the teams should be composed of both biologist and engineers. That way, the engineers will try to develop new systems in a bottom-up fashion and run numerical simulations, while the biologists will be able to assess the feasibility of such systems, and construct them from biological parts.
| + | /*********************************************** |
| | + | * Drop Down Tabs Menu- (c) Dynamic Drive DHTML code library (www.dynamicdrive.com) |
| | + | * This notice MUST stay intact for legal use |
| | + | * Visit Dynamic Drive at http://www.dynamicdrive.com/ for full source code |
| | + | ***********************************************/ |
| | | | |
| - | <P><b>Instructors:</b></P>
| + | </script> |
| - | [https://2007.igem.org/User:sven Sven Panke], Joerg Stelling
| + | |
| | | | |
| - | <P><b>Students:</b></P> | + | <!-- CSS for Drop Down Tabs Menu #1 --> |
| - | [https://2007.igem.org/User:brutsche Martin Brutsche], [https://2007.igem.org/User:kdikaiou Katerina Dikaiou], [https://2007.igem.org/User:Raphael Raphael Guebeli], [https://2007.igem.org/User:hoehnels Sylke Hoehnel], [http://leemnan.spaces.live.com Nan Li], [https://2007.igem.org/User:Stefan Stefan Luzi]
| + | <link rel="stylesheet" type="text/css" href="http://christos.bergeles.net/eth_ddcolortabs.css" /> |
| | + | <div id="colortab" class="ddcolortabs"> |
| | + | <ul> |
| | + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ" title="Home" rel="dropmenu_home"><span>Home</span></a></li> |
| | + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model" title="Modeling" rel="dropmenu_modeling"><span>System Modeling</span></a></li> |
| | + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation" title="Simulations" rel="dropmenu_simulation"><span>Simulations</span></a></li> |
| | + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology" title="System Implementation" rel="dropmenu_biology"><span>System Implementation</span></a></li> |
| | + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team" title="Meet the team" rel="dropmenu_meettheteam"><span>Meet the team</span></a></li> |
| | + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Pictures" title="Pictures!" rel="dropmenu_pictures"><span>Pictures!</span></a></li> |
| | + | </ul> |
| | + | </div> |
| | + | <div class="ddcolortabsline"> </div> |
| | | | |
| - | <P><b>Graduate Students:</b></P>
| |
| - | [http://christos.bergeles.net Christos Bergeles], [http://www.tik.ee.ethz.ch/~sop/people/thohm/ Tim Hohm], [http://fm-eth.ethz.ch/eth/peoplefinder/FMPro?-db=phonebook.fp5&-format=pf%5fdetail%5fde.html&-lay=html&-op=cn&Typ=Staff&Suche%5fText=kemmer&Suche%5fText%5fpre=kemmer&-recid=3772770936&-find=/ Christian Kemmer], [https://2007.igem.org/User:JoeKnight Joseph Knight], [https://2007.igem.org/User:uhrm Markus Uhr], [http://www.ricomoeckel.de Rico Möckel]
| |
| | | | |
| - | == Introduction to Synthetic Biology == | + | <!--1st drop down menu --> |
| - | In order to get an initial understanding of the concepts of synthetic biology, we read and presented
| + | <div id="dropmenu_home" class="dropmenudiv_a"> |
| - | publications on various topics. A representative list of the topics that we covered on this “boot-camp” is listed in the following:
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Introduction">Introduction</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Team_Members">Team Members</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Acknowledgments">Acknowledgments</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Site_Map">Site map</a> |
| | + | </div> |
| | | | |
| - | * Introduction to synthetic biology
| |
| - | * DNA de novo design
| |
| - | * DNA circuits
| |
| - | * Hysteresis
| |
| - | * Oscillators
| |
| - | * Zinc fingers
| |
| - | * Noise in single cell measurements
| |
| - | * Distance communication
| |
| - | * Parameter manipulations
| |
| - | * Protein logic
| |
| - | * Orthogonal systems
| |
| - | * mRNA engineering
| |
| - | * RNA regulators
| |
| | | | |
| - | == Choosing the Project == | + | <!--2nd drop down menu --> |
| - | === Step 1: Brainstorming === | + | <div id="dropmenu_modeling" class="dropmenudiv_a" style="width: 150px;"> |
| - | Initially, we wanted to come up with as many ideas as possible, in order to be able to choose among them the best, and find a cool project to carry out. For this reason, we had brainstorming sessions. We split up in three teams, and each team tried to come up with many fancy and showy ideas, which was facilitated by keeping in mind the following brainstorming guidelines:
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Introduction">Introduction</a> |
| - | # Defer judgment - the rules of nature don't apply | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Model_Overview">Model Overview</a> |
| - | # Encourage wild ideas
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Detailed_Model">Detailed Model</a> |
| - | # Build on the ideas of others | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Final_Model">Final Model</a> |
| - | # Be visual
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Modeling_Basics">Modeling Basics Page</a> |
| - | # Go for quantity
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Mathematical_Model">Mathematical Model</a> |
| - | # Stay focused on topic
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FSM">FSM View Page</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FlipFlop">Flip-Flop View Page</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Parameters">Parameters Page</a> |
| | + | </div> |
| | | | |
| - | The ideas that we came up with, as were presented in the following meetings, were:
| + | <!--3rd drop down menu --> |
| - | * '''PID Controller''': Design a PID controller out of biological elements. The P component can be a simple output to a regulatory protein, and the I component can be the overall protein production at a time period. What can the D component be?
| + | <div id="dropmenu_simulation" class="dropmenudiv_a" style="width: 150px;"> |
| - | * '''Motion Detector''': Cells are grown on a petri dish. Below the dish, moving images are displayed. A 3-state automaton is proposed. Output A is created when light is present. Output B is created when light is absent. Moving patterns will cause some cells to create both outputs over time. This will result in some “inspector” cells producing output C, by collecting outputs A and B.
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Introduction">Introduction</a> |
| - | * '''Analog-to-Digital Converter''': Compare the level of protein concentration with thresholds, and digitize the output.
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Simulation_of_Test_Cases">Test Cases</a> |
| - | * '''Neural Network''': Create a sort of biological neural network with bacteria. We should address the issue of learning, and find a way to incorporate the feedback in the cell decision making process. Directed evolution can be a sort of feedback, but we want to avoid this. (This idea was the basis for the “learning project”)
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Sensitivity_Analysis">Sensitivity Analysis</a> |
| - | * '''Paramedic Cells''': Some cells are able to detect signals coming from other cells, and create food for them, or create proteins in order to save them and make them function better.
| + | </div> |
| - | * '''Cell Batteries''': Cells are able to create and store large quantities of ATP, during a “storing process”. Afterwards, they can detect a signal and give back all the energy they stored, in a short burst, like a capacitor. Other ideas are that the cells can “blow up” and emit large amounts of GFP, based on the ATP that they have accumulated.
| + | |
| - | * '''Flashing Bacteria''': Cells are grown on a light pattern. The cells that are on the bright parts of the image are oscillating in phase, while the others are remaining dark. This results in the observation of a flashing pattern.
| + | |
| - | * '''Biocam''': Visible to Fluorescent light converter.
| + | |
| - | * '''BioCD''': “Print” cells on a film, then read them out and “reconstruct” the original data. Basically, it is an analog to digital converter, followed by a system that can interpret the digitized data. (This idea was the basis for the “Music of life project”, where cells would produce fluorescent proteins based on an analog input. Then, the amount and type of fluorescence would code some music).
| + | |
| - | * '''Clock''': A follow-the-leader system. We have to groups of cells. The first group creates something that repels the second group. The second group creates a protein that attracts the first group. This way, they first group wants to “catch” the second group, whereas the second group wants to “avoid” the first group. This results in them moving around. We can say that the second group is the leader, and the first group exhibits a "follow-the-leader” behavior.
| + | |
| - | * '''Sensors''': Various systems that can sense PH, pressure, temperature, meat quality, moisture e.t.c. have been proposed.
| + | |
| | | | |
| - | === Step 2: Preferred Projects === | + | <!--4th drop down menu --> |
| | + | <div id="dropmenu_biology" class="dropmenudiv_a" style="width: 150px;"> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#Introduction">Introduction</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#The_Complete_System">The Complete System</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#System_Phases">System Phases</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#Current_Cloning_Status">Current Cloning Status</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/parts">System Parts Page</a> |
| | + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/Lab">Lab Notes Page</a> |
| | + | </div> |
| | | | |
| - | We decided that three of the above ideas were worth a deeper examination. Namely, we split again in three
| + | <!--5th drop down menu --> |
| - | teams, so that we can come up with an initial system, with remarks on its feasibility and coolness. Our results
| + | <div id="dropmenu_meettheteam" class="dropmenudiv_a" style="width: 150px;"> |
| - | were presented on all the team members, so that we could then limit the potential projects down to two and
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#The_ETH_Zurich_07_Team">The ETH Zurich 07 Team</a> |
| - | one, and proceed with constructing the DNA sequence. The three projects and the presentations that we did
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#Team_Description">Team Description</a> |
| - | were:
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Internal">Brainstorming Page</a> |
| - | # '''Music of Life''': The basic idea is that instead of having an analog-to-digital converter with four outputs (three fluorescent proteins, and no output), we can have two switches. When switch A is on, RFP is produced. When switch B is on, GFP is produced. When both switches A and B are on, a yellowish output is observed. By recording these outputs, we can later create music, by assigning each fluorescent protein to a chord. For example, RFP would correspond to a G chord. The strength of the fluorescence can signify the strength of the chord. If the cells are placed on a spinning disk, we can have something like a vinyl player. A camera is observing the cells, and music is created on the fly.
| + | </div> |
| - | # '''Learning''': Based on the idea of the neural network, we want to create a biological system, where the cells can learn a specific behavior. In order to simplify the system, we decided that the cells can learn to recognize a specific type of other cells. We divide the process in a learning phase, and a recognition phase. First, cells A are put together with cells B. Then, cells A are “learning” to recognize cells B. If afterwards they are put in a petri dish with cells B, they will emit GPF. Otherwise, they will stay dark.
| + | |
| | | | |
| - | Idea two, even though it was fancy, was fairly simple, as, creating two switches is a straightforward and well understood process. As a result, it was discarded. We could try to make the system more complex, but making something more complex just for the sake of it, is not a good engineering approach ;)
| + | <script type="text/javascript"> |
| | + | //SYNTAX: tabdropdown.init("menu_id", [integer OR "auto"]) |
| | + | tabdropdown.init("colortab", 3) |
| | + | </script> |
| | | | |
| - | == The Project ==
| + | </html> |
| | | | |
| - | We decided to proceed with the '''Learning Project'''. Our goal is to design an E.coli strain, able to distinguish between two chemical substances after an assigned learning process, induced by an external chemical signal (teaching substance). After the bacterial strain was taken to a testing phase, the output will result in either yellow or cyan fluorescence, depending on whether the bacteria recognized the same chemical substance in the testing phase as it was trained in the learning phase or a different one.
| + | __NOTOC__ |
| - | We aim to implement our system using a toggle switch consisting of different repressor and activator proteins coupled with double regulatory regions.
| + | |
| | | | |
| - | Our biological system was inspired from the toggle of [?], and its binary logic equivalent. In order to understand that, one should think of learning as a "switching of behavior". As a response to an external stimulus, the behavior changes. If we approach the problem with the tools of binary logic, we can see that we want to implement a JK flip-flop with a latch. At the learning process, the JK flip-flop will stabilize at its final state Q. At the recognition phase, the flip-flop is latched, and recognition is performed (see figure below):
| + | <html> |
| | + | <p align="center"><font size="6"><b>ETH Zurich - educatETH <i>E.coli</i> System</b></font></p> |
| | + | </body> |
| | + | </html> |
| | | | |
| - | [[Image:logic_circuit.png|center]]
| + | =Introduction= |
| | + | <blockquote>"All <i>E.coli</i> 's are equal, but some <i>E.coli</i> 's are more equal than others..." ''(freely adapted from "Animal Farm" by George Orwell)''</blockquote> |
| | | | |
| - | Based on the toggle from [?], we discussed our prototype system (see figure below), which the resulted in the refined system that we describe in the next section.
| + | ... this is what George Orwell would have written, were he a synthetic biologist. In the <i>E.coli</i> colonies on petri dishes, all bacteria are equal; except for some special ones. Our project is about designing such special <i>E.coli</i> that are "more equal" than the rest: they have the ability to be trained in order to memorize and recognize their environment in the future. Their story will be presented through this wiki ... |
| | | | |
| - | (picture of initial draft of the bio system, as taken from Joe, --here--)
| + | ==Motivation== |
| | | | |
| - | === System Models ===
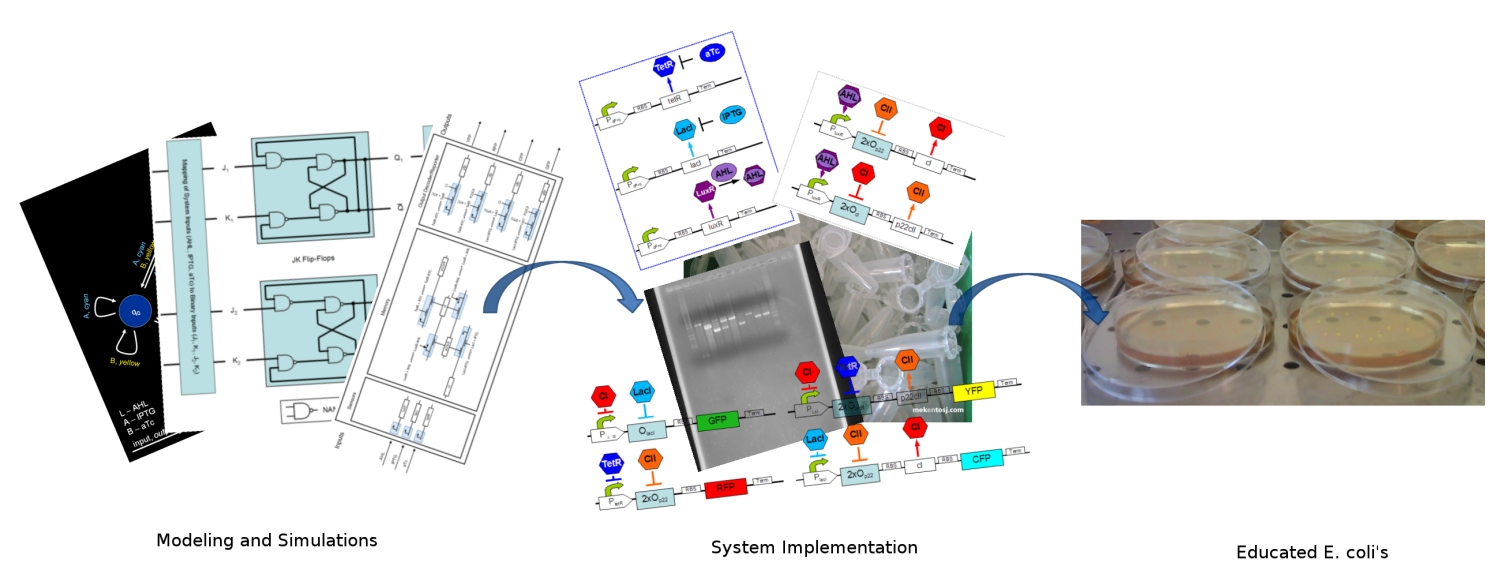
| + | [[Image:ethz_main_pic.jpg|right|thumb|<b>Fig. 1</b>: Artist's approach to the different stages of the development. We started by modeling and simulating the system. We continued by specifying the DNA strands for its implementation. Finally in the end, our system should report with different fluorescent proteins (image edited)|600px]] |
| - | [[Image:new_learning_system.jpg|center]] | + | |
| | | | |
| - | === Simulations ===
| + | Our combined team of biologists and engineers is coping with the problem of implementing memory capabilities in bacterial colonies. First, ''E.coli'' are able to respond differently (with distinct fluorescent proteins) to two different inputs (we used chemicals). Second, they remember which input was presented to them. Finally, when confronted with a new input, they are able to recognize whether it is the one that they were trained with or not. |
| - | [[ETHZ/Simulations| detailed information on the simulations]]<BR>
| + | In other words, in this project '''we are educating the <i>E.coli</i>'''! |
| | | | |
| - | === Sensitivity Analysis === | + | ===Multipurpose Cell Lines=== |
| | | | |
| - | == Lab Work ==
| + | Our system has the ability to behave in different ways according to an internal toggle inside it switching states based on the chemical substances that the system is exposed to. The toggle states could generally be used to trigger events such as enzyme synthesis, transcriptional regulation, virion production, or even cell death. Therefore, one may view the bacterial cell line containing this system as a multipurpose cell line. By adding a certain chemical to a cell line, the latter may be trained to exhibit a desired behavior, and then it is not necessary any more to construct two independent cell lines. |
| | | | |
| - | Here we can write when each one will be available. For example:
| + | This means that one applies an “input engineering” instead of a “DNA engineering” approach. If one extends this idea to several multi-inducible toggle switches being harbored in the same cell line, the number of possible phenotypes increases to 2<i><sup>n</sup></i>, where <i>n</i> equals the number of toggle switches. For example, if one would have 5 such toggle switches inside a cell line, 32 different behavior patterns would be possible. |
| - | *<b>Christos:</b><br> Can be available everyday after 18:00, till 00:00. Weekends as well.
| + | |
| - | *<b>Tim:</b><br> 13.08.-15.08. after 17:30<br> away from 17.08. until 02.09.
| + | |
| - | *<b>Martin:</b><br> Every day, also weekends, for half a day or even more if it's requiered. The last week of August and the first one in September I have only time for aproximately 5-6 h a day due to learning for the exams.
| + | |
| - | *<b>Katerina:</b><br> Not available 5.09-7.09 (final exams). Otherwise available after 17:30. Possible also in the morning, if I know in advance that I will be needed.
| + | |
| - | *<b>Christian:</b><br> Not available 5.10-24.10.2007 --> holidays, I can do most of the labwork during the day in my lab
| + | |
| | | | |
| - | === Cloning Plan ===
| + | For the purpose of creating a toggle switch that is activated in a specific phase only and not always (a multi-inducible toggle switch), as is required for stable biological automatons, we introduced the concept of [https://2007.igem.org/ETHZ/Biology/parts#double_promoters double promoters] to the [http://partsregistry.org/Main_Page Registry of Standard Biological Parts], which can be helpful for engineering systems which exhibit a desired behavior only at specific times. |
| | | | |
| - | The system consists of 11 parts. These are:
| + | ===Link to Epigenetics=== |
| | + | Epigenetics refers to features like chromatin or DNA modifications that do not involve changes in the underlying DNA sequence and are stable over many cell divisions [1],[2]. If one has a closer look at our proposed system, one can also view it as a model-system for epigenetics: Although the DNA sequence itself stays the same, two different subpopulations of cells with different phenotypes can develop from it. Put simply, depending in which state (subpopulation) the toggle switch is, the cells will produce different fluorescent proteins upon addition of two different inducer molecules. Therefore, the epigenetic feature here is the binding of specific repressor proteins whose production is dependent on the toggle switch state. |
| | | | |
| - | {| class="wikitable" border = "1" style="text-align:left"
| + | ===Intelligent Biosensors and Self-Adaptation=== |
| - | |+ '''System parts'''
| + | |
| - | |-
| + | |
| | | | |
| | + | The system is capable of sensing different chemicals and producing different fluorescent proteins. Since the cells can be trained to produce one of several specific fluorescent protein types when a certain chemical is present, one can also view those cells as intelligent biosensors which recognize chemical substances according to a training phase. The intelligent biosensors are not limited to detect chemicals; temperature, pH, light, pressure could be detected with an appropriate system as well. Such an application could be especially of interest when the environment to be probed is harmful for humans, for example due to high toxicity. |
| | | | |
| | + | ==Team Members== |
| | + | [[Image:ETHZ_Group_photo_6.png|right|thumb|The ETH Zurich iGEM2007 Team|270px]] |
| | | | |
| - | |-
| + | The ETH Zurich team consists of a good mixture between biologists and engineering students. |
| | + | We are: |
| | + | <ul> |
| | + | <li><i>Undergraduate students</i>: <br> |
| | + | [https://2007.igem.org/User:brutsche Martin Brutsche], [https://2007.igem.org/User:kdikaiou Katerina Dikaiou], <br>[https://2007.igem.org/User:Raphael Raphael Guebeli], [https://2007.igem.org/User:hoehnels Sylke Hoehnel], <br> |
| | + | [https://2007.igem.org/Nan_Li Nan Li], |
| | + | [https://2007.igem.org/User:Stefan Stefan Luzi] |
| | + | <li><i>Graduate students</i>: <br> |
| | + | [http://christos.bergeles.net Christos Bergeles], |
| | + | [http://www.tik.ee.ethz.ch/~sop/people/thohm/ Tim Hohm], <br> |
| | + | [http://www.fussenegger.ethz.ch/people/kemmerc Christian Kemmer], |
| | + | [https://2007.igem.org/User:JoeKnight Joseph Knight], <br> |
| | + | [http://www.ricomoeckel.de Rico Möckel], |
| | + | [http://csb.inf.ethz.ch/people/uhr.html Markus Uhr] |
| | + | <li><i>Project advisors</i>: <br> |
| | + | [http://www.ipe.ethz.ch/laboratories/bpl/people/panke Sven Panke], <br> |
| | + | [http://csb.inf.ethz.ch/people/stelling.html Joerg Stelling] |
| | + | </ul> |
| | | | |
| - | ! 1
| + | For more information about us, visit our [[ETHZ/Meet_the_team | Meet the Team]] page. |
| | | | |
| - | | TetR production(constitutive part of system)
| + | ==Acknowledgments== |
| | + | The idea for the project as well as its implementation was done solely by the ETH iGEM 2007 team. We would like to thank the people in [http://www.ipe.ethz.ch/laboratories/bpl/index Sven Panke's Lab], especially Andreas Meyer who was always there for us when we had a problem. Additionally, we would like to thank [http://www.facs.ethz.ch Alfredo Franco-Obregón's lab] and Oralea Büchi for the help with the flow cytometry. |
| | | | |
| | + | We would also like to acknowledge the financial support by [http://europa.eu EU], the [http://www.ethz.ch ETH Zurich], and [http://www.geneart.com GeneArt]: |
| | + | <center> |
| | + | {| border="0" |
| | |- | | |- |
| | + | | [http://europa.eu http://www.tik.ee.ethz.ch/~thohm/EU.gif] |
| | + | | width="40" | |
| | + | | [http://www.ethz.ch http://www.tik.ee.ethz.ch/~thohm/ethlogo.jpg] |
| | + | | width="40" | |
| | + | | [http://www.geneart.com http://www.tik.ee.ethz.ch/~thohm/geneart.gif] |
| | + | |} |
| | + | </center> |
| | | | |
| - | ! 2
| + | ==Site Map== |
| | | | |
| - | | LacI production (constitutive part of system) | + | In this wiki, we will present to you a detailed description of the proposed system: starting with the [[ETHZ/Model | modeling of the system]], we describe both, [[ETHZ/Simulation | simulations and theoretical considerations]] of the system, as well as the actual [[ETHZ/Biology | implementation using bio-bricks]] accompanied by our [[ETHZ/Biology/Lab | lab notes]]. Additionally, you will find further [[ETHZ/Meet_the_team | information on the team]], more details about [[ETHZ/Internal | ideas we developed]] before we came up with the system we finally implemented, and some [[ETHZ/Pictures | pictures]] documenting our work. |
| | | | |
| - | |-
| + | The site map of our wiki is the following: |
| | | | |
| - | ! 3 | + | {| class="wikitable" width="100%" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" |
| - | | + | !width="34%"| Modeling Pages |
| - | | LuxR production(constitutive part of system) | + | !width="22%"| Biology Pages |
| | + | !width="22%"| ETHZ Team Pages |
| | + | !width="22%"| Links |
| | |- | | |- |
| - | | + | | [[ETHZ/Model | Modeling of the learning system]] |
| - | ! 4
| + | | [[ETHZ/Biology | Biological implementation]] |
| - | | + | | [[ETHZ/Meet_the_team | Team page]] |
| - | | 1st half of p22/YFP production (outer part of system, reporting) | + | | [https://2006.igem.org/wiki/index.php/ETH_Zurich_2005 The ETH Zurich 2005 project] |
| | |- | | |- |
| - | | + | | [[ETHZ/FlipFlop | Representation using flip-flops]] |
| - | ! 5
| + | | [[ETHZ/Biology/parts | Biobricks/parts]] |
| - | | + | | [[ETHZ/Pictures | Pictures]] |
| - | | 2nd half of p22/YFP production (outer part of system, reporting) | + | | [https://2006.igem.org/wiki/index.php/ETH_Zurich_2006 The ETH Zurich 2006 project] |
| | |- | | |- |
| - | | + | | [[ETHZ/FSM | Representation using finite state machines]] |
| - | !6
| + | | [[ETHZ/Biology/Lab | Lab notes]] |
| - | | + | | [[ETHZ/Internal | Brainstorming sessions]] |
| - | | CI production (inner part of system) | + | | |
| | |- | | |- |
| - | | + | | [[ETHZ/Simulation | Model simulations and theoretical considerations]] |
| - | ! 7
| + | | |
| - | | + | | |
| - | | p22 production (inner part of system) | + | | |
| | |- | | |- |
| - | | + | | [[ETHZ/Parameters | Parameters used in our simulations]] |
| - | ! 8
| + | | |
| - | | + | | |
| - | | 1st half of CI/CFP production (outer part of system, reporting) | + | | |
| | |- | | |- |
| - |
| |
| - | ! 9
| |
| - |
| |
| - | | 2nd half of CI/CFP production (outer part of system, reporting)
| |
| - | |-
| |
| - |
| |
| - | ! 10
| |
| - |
| |
| - | | RFP production (reporting)
| |
| - | |-
| |
| - |
| |
| - | ! 11
| |
| - |
| |
| - | | GFP production (reporting)
| |
| - |
| |
| | |} | | |} |
| | | | |
| - | 3 plasmids will be used for the 11 system parts as follows:
| + | == References == |
| - | | + | |
| - | {| class="wikitable" border = "1" style="text-align:left"
| + | |
| - | |+'''Plasmids and contents'''
| + | |
| - | |-
| + | |
| - | ! plasmid !! resistance !! copy type!! contents !! comments
| + | |
| - | |-
| + | |
| - | | + | |
| - | | [[ETHZ/pbr322| pbr322]] || ampicillin || medium || 1,2,3 || constitutive part
| + | |
| - | |-
| + | |
| - | | + | |
| - | | [[ETHZ/pck01| pck01]] || chloramphenicol|| low || 4,5,8,9 || outer part
| + | |
| - | |-
| + | |
| - | | + | |
| - | | [[ETHZ/pacyc177| pacyc177]] || kanamycin|| low || 6,7,10,11 || inner part, reporting
| + | |
| - | |-
| + | |
| - | | + | |
| - | |}
| + | |
| - | | + | |
| - | The standard BioBrick assembly will be used to put the parts in the plasmids. Detailed information on how the BioBrick part fabrication works can be found [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication here]. For a shorter explanation of how to assemble 2 parts together check [http://partsregistry.org/Assembly:Standard_assembly here]. Note that the composite part is constructed from the end to the beginning, i.e. each new part is inserted ''before'' the existing one. In the following, the plasmid containing the new part to be inserted will be referred to as the ''donor'' and the plasmid accepting the new part will be referred to as the ''acceptor''. Composite pars made of parts '''a''' and '''b''' are denoted '''a.b'''.
| + | |
| - | | + | |
| - | ==== '''Plasmid 1 ''(pbr322ap)'' ''' ==== | + | |
| - | | + | |
| - | # Put parts 1,2,3 in pbr322ap plasmids.
| + | |
| - | # Merge plasmid containing part '''2''' ''(donor)'' with plasmid containing part '''3''' ''(acceptor)''. You should get a plasmid containing a '''2.3''' composite part.
| + | |
| - | # Merge plasmid containing part '''1''' ''(donor)'' with plasmid containing composite part '''2.3''' ''(acceptor)''. You should get a plasmid containing a '''1.2.3''' composite part.
| + | |
| - | | + | |
| - | ==== '''Plasmid 2 ''(pck01cm)'' ''' ====
| + | |
| - | | + | |
| - | # Put parts 4,5,8,9 in pck01cm plasmids.
| + | |
| - | # Merge plasmid containing part '''4''' ''(donor)'' with plasmid containing part '''5''' ''(acceptor)''. You should get a plasmid containing a '''4.5''' composite part.
| + | |
| - | # Merge plasmid containing part '''8''' ''(donor)'' with plasmid containing part '''9''' ''(acceptor)''. You should get a plasmid containing a '''8.9''' composite part. ''Note'': this step can be done simultaneously with the above.
| + | |
| - | # Merge plasmid containing composite part '''4.5''' ''(donor)'' with plasmid containing composite part '''8.9''' ''(acceptor)''. You should get a plasmid containing a '''4.5.8.9''' composite part.
| + | |
| - | | + | |
| - | ==== '''Plasmid 3 ''(pacyc177km)'' ''' ====
| + | |
| - | | + | |
| - | # Put parts 6,7,10,11 in pacyc177km plasmids.
| + | |
| - | # Merge plasmid containing part '''6''' ''(donor)'' with plasmid containing part '''7''' ''(acceptor)''. You should get a plasmid containing a '''6.7''' composite part.
| + | |
| - | # Merge plasmid containing part '''10''' ''(donor)'' with plasmid containing part '''11''' ''(acceptor)''. You should get a plasmid containing a '''10.11''' composite part. ''Note'': this step can be done simultaneously with the above.
| + | |
| - | # Merge plasmid containing composite part '''6.7''' ''(donor)'' with plasmid containing composite part '''10.11''' ''(acceptor)''. You should get a plasmid containing a '''6.7.10.11''' composite part.
| + | |
| - | | + | |
| - | | + | |
| - | ==== '''Linkers''' ====
| + | |
| - | | + | |
| - | # [[ETHZ/pCK01-1| pck01-1]]
| + | |
| - | # [[ETHZ/pCK01-2| pck01-2]]
| + | |
| - | # [[ETHZ/pacyc177-1| pacyc177-1]]
| + | |
| - | # [[ETHZ/pacyc177-2| pacyc177-2]]
| + | |
| - | # [[ETHZ/pbr322-1| pbr322-1]]
| + | |
| - | # [[ETHZ/pbr322-2| pbr322-2]]
| + | |
| - | # [[ETHZ/pbr322-3| pbr322-3]]
| + | |
| - | # [[ETHZ/pbr322-4| pbr322-4]]
| + | |
| - | | + | |
| - | We have got 4 linkers for pbr322, because there are two for the tetracycline version of pbr333 and two for the ampicillin version.
| + | |
| - | | + | |
| - | === Lab Instructions and Methods ===
| + | |
| - | | + | |
| - | === Lab Schedule ===
| + | |
| - | <font face="DejaVu Sans" size="3"><b>Week 1</b></font> <b>06.08.07 - 12.08.07</b><br>
| + | |
| - | [[ETHZ/labnotes| lab schedule week 1]]<BR>
| + | |
| - | <font face="DejaVu Sans" size="3"><b>Week 2</b></font> <b>13.08.07 - 19.08.07</b><br>
| + | |
| - | [[ETHZ/labnotes| lab schedule week 2]]<BR>
| + | |
| - | <font face="DejaVu Sans" size="3"><b>Week 3</b></font> <b>20.08.07 - 26.08.07</b><br>
| + | |
| - | [[ETHZ/labnotes| lab schedule week 3]]<BR>
| + | |
| - | <font face="DejaVu Sans" size="3"><b>Week 4</b></font> <b>27.08.07 - 02.09.07</b><br>
| + | |
| - | [[ETHZ/labnotes| lab schedule week 4]]<BR>
| + | |
| - | <font face="DejaVu Sans" size="3"><b>Week 5</b></font> <b>03.08.07 - 09.09.07</b><br>
| + | |
| - | [[ETHZ/labnotes| lab schedule week 5]]<BR>
| + | |
| - | <font face="DejaVu Sans" size="3"><b>Week 6</b></font> <b>10.09.07 - 16.09.07</b><br>
| + | |
| - | [[ETHZ/labnotes| lab schedule week 6]]<BR>
| + | |
| - | <font face="DejaVu Sans" size="3"><b>Week 7</b></font> <b>17.09.07 - 23.09.07</b><br>
| + | |
| - | [[ETHZ/labnotes| lab schedule week 7]]<BR>
| + | |
| - | <font face="DejaVu Sans" size="3"><b>Week 8</b></font> <b>24.09.07 - 30.09.07</b><br>
| + | |
| - | [[ETHZ/labnotes| lab schedule week 8]]<BR>
| + | |
| - | <font face="DejaVu Sans" size="3"><b>Week 9</b></font> <b>01.10.07 - 07.10.07</b><br>
| + | |
| - | [[ETHZ/labnotes| lab schedule week 9]]<BR>
| + | |
| - | | + | |
| - | === Lab Notes / Log Book ===
| + | |
| - | | + | |
| - | Here we can write down every day a detailed version of the work we have done (it should be the same stuff we write in the "real lab book" which stays in the lab). In this way everyone can see in detail what was done in the lab.
| + | |
| | | | |
| - | [[ETHZ/Lab book| detailed lab notes]]<BR> | + | [http://www.nature.com/nature/journal/v447/n7143/abs/nature05913.html;jsessionid=62903C604764B175945C03DB8639ECBD [1] Bird A] <i>"Perceptions of epigenetics"</i>, Nature 447:396-398, 2007 <br /> |
| | + | [http://linkinghub.elsevier.com/retrieve/pii/S096098220701007X [2] Ptashne M] <i>"On the use of the word ‘epigenetic’"</i>, Current Biology 17(7):R233-R236, 2007 <br /> |
| | | | |
| - | == Task List ==
| |
| | | | |
| - | The things to do, from the most pressing (timewise), to the least pressing (timewise) is below. Please put your name next to the task that you believe that you can undertake.
| |
| | | | |
| - | # '''Team descriptions (overdue) and team photograph''' <br> ''Christian'': I guess I am the only semiprofessional photographer of the group. I can do some group pictures etc. but for this we need some ideas... - I would like to do something special. I also made the group-pics of the Synth. Biology 3.0 conference. Some references ;-) : [http://www.fotocommunity.de/pc/pc/mypics/461397] <br> ''Raphael'': What about a short movie of us?
| + | <html><body> |
| - | # '''Team rosters due (1.9.)'''<br> ''Martin'': Does somebody know, what exactly should be done here? <br> ''Nan'': A list of team members, including some basic personal info.? (e.g. nationality, background, pet peeves...? )<br>''Katerina'': Guys, to make this easier, either write things about you on your personal page on the wiki or link to a page about you. This way it'll be easy to put it all together afterwards.
| + | <a href="http://www3.clustrmaps.com/counter/maps.php?url=https://2007.igem.org/ETHZ" id="clustrMapsLink"><img src="http://www3.clustrmaps.com/counter/index2.php?url=https://2007.igem.org/ETHZ" style="border:0px;" alt="Locations of visitors to this page" title="Locations of visitors to this page" id="clustrMapsImg" onError="this.onError=null; this.src='http://www2.clustrmaps.com/images/clustrmaps-back-soon.jpg'; document.getElementById('clustrMapsLink').href='http://www2.clustrmaps.com'" /> |
| - | # '''Labwork (parts have to be at the registry in Boston on 26.10.)''' <br> ''Joe'': I can be in the lab at least 2 evenings a week and some times through the weekdays. <br> ''Martin'': From Monday I can work every day for the whole day. At the moment I only work for several hours... <br> ''Rico'': I have my exam on Tuesday. Afterwards I can assist.<br> ''Christian'': I can do the introduction of the polylinker into the vectors beside my normal labwork on the Hoenggerberg. I could also do the whole biobrick assembly if you want this (I will go on holiday from the 5.10-24.10.07) <br> ''Raphael'': That's the part where I will mainly contribute, from 14.09. on I can work several days/week<br> ''Christos'': I can assist at the afternoons, if needed. <br> ''Katerina'': 7.09 - 30.09 generally plenty of time, apart from when I do my semester project presentation, will keep you posted when that is.
| + | </a> |
| - | # '''Simulation and sensitivity analysis'''<br> ''Martin'': From 10. Sept. I've got plenty time to work on it. I think Markus would join here too ;-) <br> ''Tim'': I can contribute in running stuff and help identify parameters from literature <br> ''Rico'': I can do simulations, sensitivity analysis. <br> ''Nan'': I would like to do simulations and sensitivity analysis. <br> ''Christos'': Yeap, I guess I can be here too. I will check some toolboxes to automate things, this weekend. <br> ''Katerina'': Want to help (parameter identification, programming, sensitivity analysis). Discuss in upcoming meeting tasks, versioning and ask Christian about parameters.
| + | |
| - | # '''Presentation''' <br> ''Joe'': I'm American... I can sale anything.<br> ''Martin'': I'm bad in Layout stuff, but maybe I can help as an idea supplier or so. <br> ''Rico'': I like giving presentations. For preparation we will need a mixture of different background and excellent pictures!!! <br> ''Christian'': I can provide the molecular biology knowledge and part. <br> ''Nan'': I can work on the slides. <br> ''Christos'': I like this part, I guess everyone will contribute anyway...<br> ''Katerina'': I believe I can be of help in structure, layout and fancy stuff. Could help train a bit the people we decide to do the presentation (question answering, style). I also think that Joe and Christos could be good for doing the presentation (structured, pleasant voices and lively), can discuss this in a meeting.
| + | <html><body> |
| - | #''' Poster''' <br> ''Joe'': See 5. above <br> ''Rico'': I can help. <br> ''Christian'': I can provide the molecular biology knowledge and part. <br> ''Nan'': Partly art. I will help. <br> ''Christos'': You can sell, but can you trick? lol :)<br> ''Katerina'': Not my strong point, but have an eye for typos, fonts, layout etc, so can help in final checking.
| + | <a> |
| - | # '''Wiki (Project and part documentation due on 26.10)''' <br> ''Martin'': See the points above, from next Monday I will give everything, now I'm doing my best... <br> ''Rico'': I can help. I guess this will have to contain the materials that we will also use for poster and presenttion anyway. <br> ''Nan'': Shouldn't it be updated with every going on process? <br> ''Christos'': I am trying to put stuff in, as it comes along. I will update the bio pages with the presentation material, this weekend (I hope). <br> ''Katerina'': You guys have done a great job so far, will help with whatever needed.
| + | <img src="http://www.easycounter.com/counter.php?stefanluzi" |
| | + | border="0" alt="Welcome to ETHz - iGEM07"></a> |
| | + | </body></html> |