Boston University
From 2007.igem.org
(→Images) |
|||
| Line 21: | Line 21: | ||
| - | {| style="width:75%; height:200px" border="1" | + | |
| + | {| align="center" style="width:75%; height:200px; color:white;" border="1" | ||
|- | |- | ||
| - | | align="center" | Our Team | + | | bgcolor="#990000" color="white" height="30pt" align="center" | '''Our Team''' |
| - | | align="center" | Project Design | + | | bgcolor="#990000" color="white" align="center" | '''Project Design''' |
| - | | align="center" |Project Results | + | | bgcolor="#990000" color="white" align="center" | '''Project Results''' |
| - | | align="center" | Miscellany | + | | bgcolor="#990000" color="white" align="center" | '''Miscellany''' |
| - | |- | + | |- style="color:#990000;" |
| - | | align="center" | jkl | + | | align="center" | '''jkl''' '''hahaha''' |
| - | | align="center" | mno | + | | align="center" | '''mno''' |
| - | | align="center" | pqr | + | | align="center" | '''pqr''' |
| - | | align="center" | stv | + | | align="center" | '''stv''' |
|- | |- | ||
|} | |} | ||
Revision as of 16:06, 6 July 2007
Contents |
About Us
Welcome to the wiki for Boston University's iGEM 2007 team!
Our team consists of David Shi, Rahul Ahuja, Christian Ling, and Danny Bellin, all soon-to-be juniors majoring in Biomedical Engineering at Boston University.
We are advised by [http://www.bu.edu/dbin/bme/faculty/?prof=tgardner Dr. Timothy Gardner], Assistant Professor of Biomedical Engineering, as well as Frank Juhn, Kevin Litcofsky, and Stephen Schneider, students in the [http://gardnerlab.bu.edu/ Gardner Laboratory], where we work. We are grateful to our advisors for their time and support!
We are also grateful to [http://www.pfizer.com Pfizer], the [http://www.bu.edu/eng Boston University College of Engineering], and the [http://www.bu.edu/eng/bme Boston University Department of Biomedical Engineering], for their generous support of our team.
Our Project
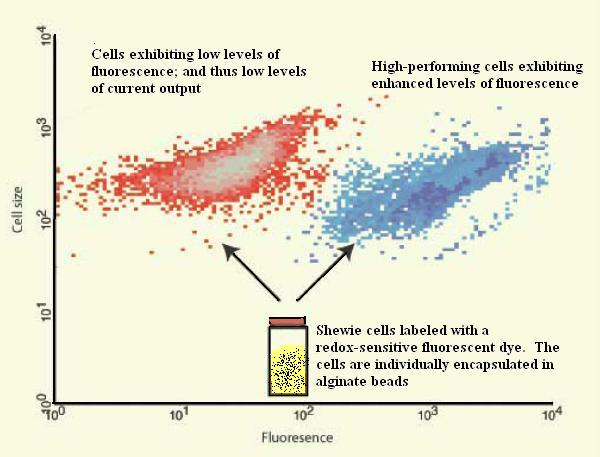
The goal of our project is to use directed evolution to increase the current output of the electrogenic bacteria Shewanella oneidensis (affectionately referred to as Shewie in the Gardner Lab). As the name suggests, directed evolution consists of two main steps: intentionally mutating DNA and then selecting for the expression of desired traits.
In the case of S. oneidensis, certain global transcription regulators in its genome have been identified as being related to the metabolic processes of the bacteria. These global transcription regulators will be mutated via error-prone PCR and transformed into S. oneidensis in hopes of altering current output. Bacteria that express greater electrogenic capability will then be selected via flow cytometry or other viable selection methods. This process of directed evolution can be repeated with previously selected S. oneidensis in order to increase the level of electrogenesis even further.
Project Progress
| Our Team | Project Design | Project Results | Miscellany |
| jkl hahaha | mno | pqr | stv |