Davidson Missouri W
From 2007.igem.org
Macampbell (Talk | contribs) |
|||
| Line 60: | Line 60: | ||
• Malcolm Campbell [http://www.bio.davidson.edu/people/macampbell/macampbell.html], Professor, Department of Biology, [mailto:macampbell@davidson.edu] | • Malcolm Campbell [http://www.bio.davidson.edu/people/macampbell/macampbell.html], Professor, Department of Biology, [mailto:macampbell@davidson.edu] | ||
| - | • | + | • [http://www.bio.davidson.edu/people/kahaynes/kahaynes.html Karmella Haynes], Visiting Assistant Professor, Department of Biology, [mailto:kahaynes@davidson.edu] |
• Laurie Heyer [http://www.davidson.edu/math/heyer/], Associate Professor, Department of Mathematics, [mailto:laheyer@davidson.edu] | • Laurie Heyer [http://www.davidson.edu/math/heyer/], Associate Professor, Department of Mathematics, [mailto:laheyer@davidson.edu] | ||
Revision as of 01:01, 17 June 2007
===Davidson & Missouri Western Team Logos, iGEM2006===
Contents |
Team Meeting Notes
Western Meeting Notes 051407 to Present [1]
Math Related Notes
Notes 051407 to Present [2]
Students
• Will DeLoache, Junior Biology Major, [3]
• Oyinade Adefuye, Senior Biology Major, [4]
• Jim Dickson, Junior Math and Economics Major, [5]
• Amber Shoecraft, Math Major, [6]
• Andrew Martens, Senior Biology Major, [7]
• Michael Waters, Sophomore Biology Major, [8]
• Jordan Baumgardner, Junior Biology, Biochemistry/Molecular Biology Major, [9]
• Ryan Chilcoat, Junior Biology Major (Health Sciences), [10]
• Tom Crowley, Senior Biochemisty/Molecular Biology Major, [11]
• Lane H. Heard, Central High School graduate, [12]
• Nickolaus Morton, Junior Chemistry Major, [13]
• Michelle Ritter, Junior Mathematics Major, [14]
• Jessica Treece, Junior Biology Major (Health Sciences), [15]
• Matthew Unzicker, Senior Biochemistry/Molecular Biology Major, [16]
• Amanda Valencia, Senior Biochem/Molecular Biology Major, [17]
Faculty
• Malcolm Campbell [http://www.bio.davidson.edu/people/macampbell/macampbell.html], Professor, Department of Biology, [18]
• [http://www.bio.davidson.edu/people/kahaynes/kahaynes.html Karmella Haynes], Visiting Assistant Professor, Department of Biology, [19]
• Laurie Heyer [http://www.davidson.edu/math/heyer/], Associate Professor, Department of Mathematics, [20]
Shipping Address: Malcolm Campbell, Biology Dept. Davidson College, 209 Ridge Road, Davidson, NC 28036 [(704) 894-2692]
• Todd Eckdahl [http://staff.missouriwestern.edu/~eckdahl/], Professor, Department of Biology, [21]
• Jeff Poet [http://staff.missouriwestern.edu/~poet/], Assistant Professor, Department of Computer Science, Mathematics, and Physics, [22]
Shipping Address: Todd Eckdahl, Biology Department, Missouri Western State University, 4525 Downs Drive, Saint Joseph, MO, 64507 [(816) 271-5873]
Project Overview
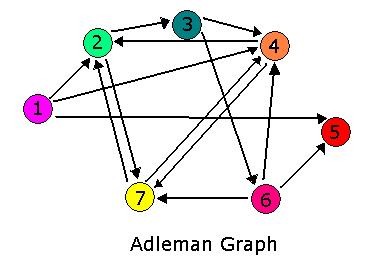
Hamiltonian Path Problem As a part of iGEM2006, a combined team from Davidson College and Missouri Western State University reconstituted a hin/hix DNA recombination mechanism which exists in nature in Salmonella as standard biobricks for use in E. coli. The purpose of the 2006 combined team was to provide a proof of concept for a bacterial computer in using this mechanism to solve a variation of The Pancake Problem from Computer Science. This task utilized both biology and mathematics students and faculty from the two institutions.
For 2007, we continue our collaboration and our efforts to manipulate E. coli into mathematics problem solvers as we refine our efforts with the hin/hix mechanism to explore another mathematics problem, the Hamiltonian Path Problem. This problem was the subject of a groundbreaking paper by Adelman in 1994 (citation below) where a unique Hamiltonian path was found in vitro for a particular directed graph on seven nodes. We propose to make progress toward solving the particular problem in vivo.
Splitting Genes for HPP
Resources / Citations
Missouri Western's Wet Lab Protocols
[http://gcat.davidson.edu/iGEM07/genesplitter.html Spliting Genes Web Tool]
Cool site for Breakfast [http://www.cut-the-knot.org/SimpleGames/Flipper.shtml]
Karen Acker's paper describing GFP and TetA(c) with Hix insertions [http://www.bio.davidson.edu/Courses/Immunology/Students/spring2006/Acker/Acker_finalpaperGFP.doc]
Bruce Henschen's paper describing one-time flippable Hix sites [http://www.bio.davidson.edu/Courses/genomics/2006/henschen/Bruce_Finalpaper.doc]
Intro to Hamiltonian Path Problem and DNA [http://www.ams.org/featurecolumn/archive/dna-abc2.html]
Adelman, LM. Molecular Computation of Solutions To Combinatorial Problems. Science. 11 November 1994. Vol. 266. no. 5187, pp. 1021 - 1024
Ptashne, Mark. A Genetic Switch: Phage Lambda Revisited, Third Edition. New York. Cold Spring Harbor Laboratory Press: 2004.
Sambrook and Russell. 2001. Molecular Cloning A Laboratory Manual. Cold Spring Harbor Laboratry Press. Cold Spring Harbor, New York pg. 1.145. 2007 June.
Literature and Registry Research
Registry Search for Possible Promoters:
BBa_J24669 --- arabinose induced
BBa_R0082 --- Is upstream of the ompC porin gene
BBa_R0074 --- Penl regulated
BBa_I14017 --- P(Rhl)
BBa_I14018 --- P (Bla) --> amp resistance
BBa_J3902 --- Pr Fe (Pl + Pll rus operon)
BBa_R0077 --- CinR --> thought to have own terminator
BBa_R0078 --- CinR (no RBS)
BBa_R0062 --- luxR & HSL regulated -- luxpR
Possibly the use of Constitutive Promoter Family Members to strengthen other promoters.
Literature Search for Polycistronic Genes on Plasmids
Sol Operon http://www.jstage.jst.go.jp/article/bbb/71/1/58/_pdf
Transfer (tra) Operon http://www.pubmedcentral.nig.gov/picrender.fcgi?artid=1347297&blobtype=pdf
Oligopeptide Permease (opp) http://www.pubmedcentral.nig.gov/picrender.fcgi?artid=1087318&blobtype=pdf