ETHZ/Biology/Lab
From 2007.igem.org
m |
|||
| Line 154: | Line 154: | ||
Ampicillin | Ampicillin | ||
| | | | ||
| - | [[Image: | + | [[Image:MapI739201.jpg|center|thumb|BBa_I739201 Map|100px]] |
|- | |- | ||
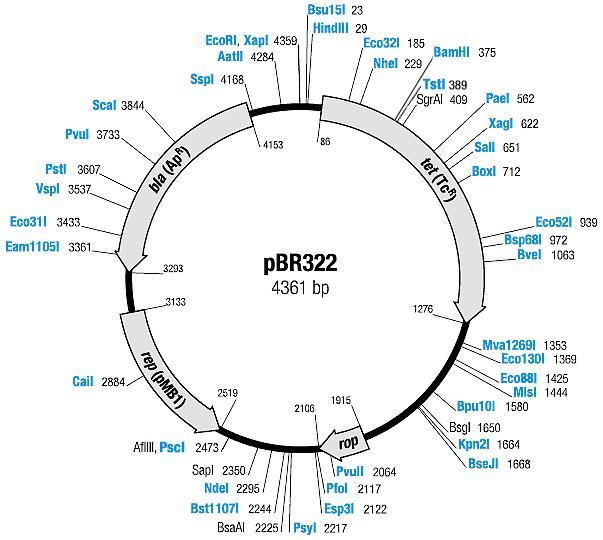
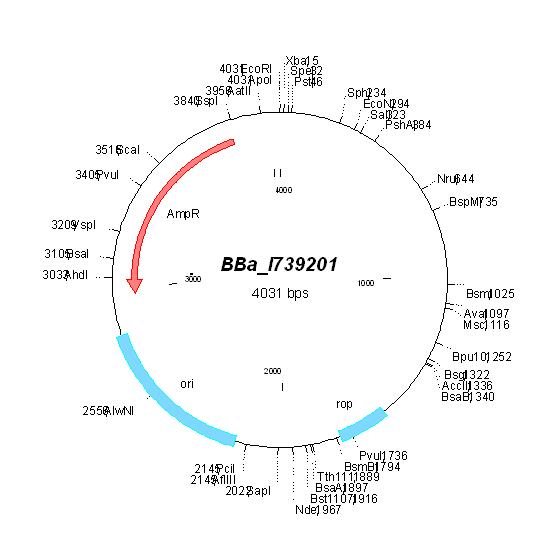
| [[ETHZ/pbr322| pBR322]] | | [[ETHZ/pbr322| pBR322]] | ||
| Line 164: | Line 164: | ||
Tetracycline | Tetracycline | ||
| | | | ||
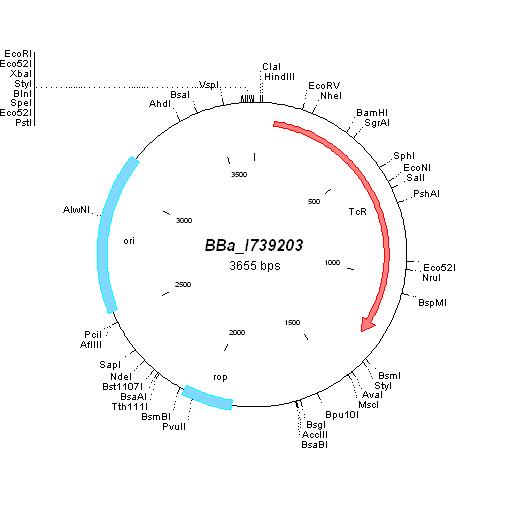
| + | [[Image:MapI739203.jpg|center|thumb|BBa_I739203 Map|100px]] | ||
|- | |- | ||
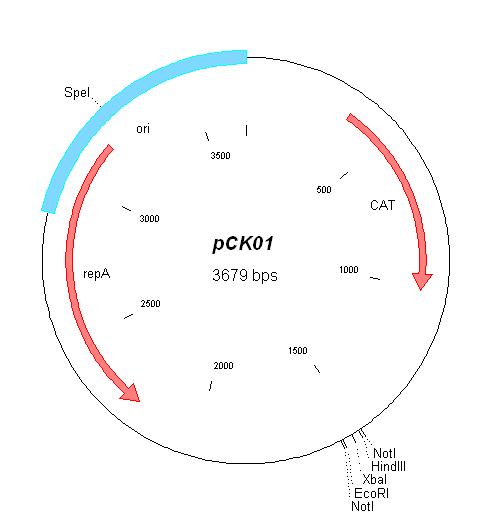
| [[ETHZ/pck01| pCK01]] | | [[ETHZ/pck01| pCK01]] | ||
| Line 174: | Line 175: | ||
Chloramphenicol | Chloramphenicol | ||
| | | | ||
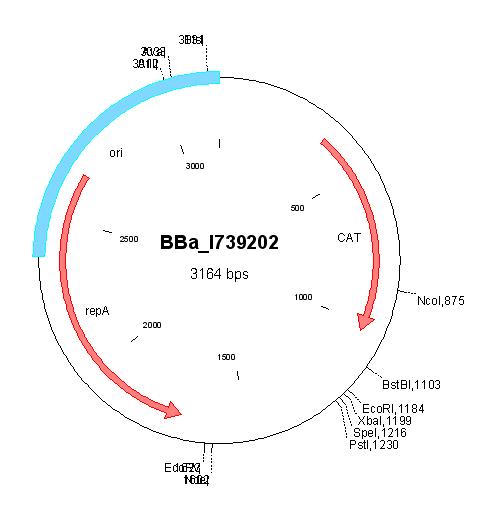
| + | [[Image:MapI739202.jpg|center|thumb|BBa_I739202 Map|100px]] | ||
|- | |- | ||
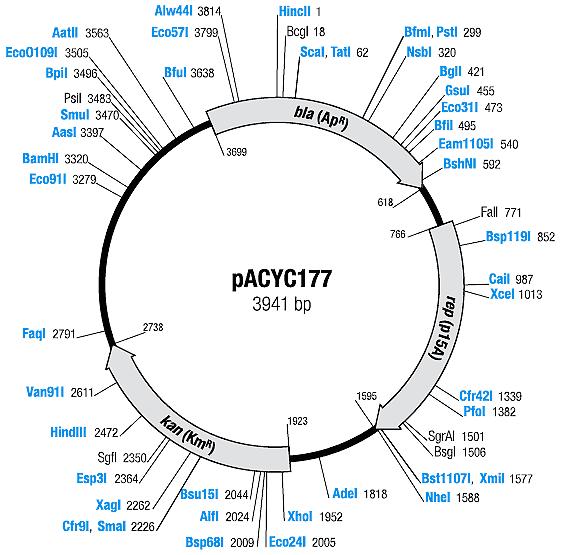
| [[ETHZ/pacyc177| pACYC177]] | | [[ETHZ/pacyc177| pACYC177]] | ||
| Line 182: | Line 184: | ||
| | | | ||
Kanamycin | Kanamycin | ||
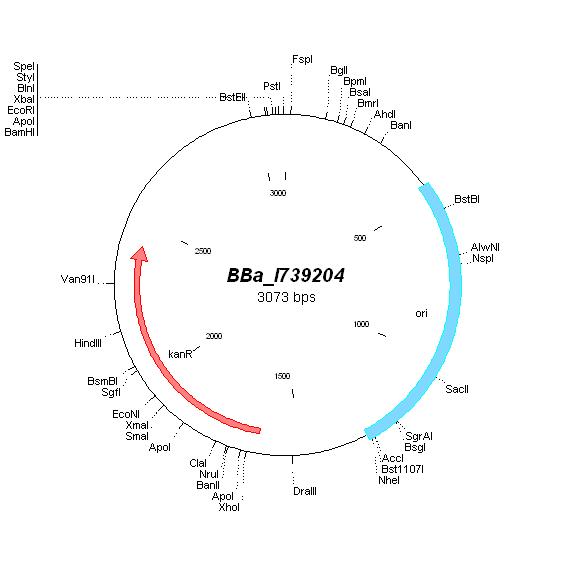
| - | | [[Image: | + | | [[Image:MapI739204.jpg|center|thumb|BBa_I739204 Map|100px]] |
|- | |- | ||
|} | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
==Cloning plan== | ==Cloning plan== | ||
Revision as of 20:39, 24 October 2007
On this page, you may find information about how educatETH e.coli was implemented in the lab. More specifically, you will find information on the plasmid strains we used, the modifications we did to them in order to be compatible with the Biobrick library and our cloning plan. Moreover, in this page you may find here an (unfortunately not complete) electronic copy of our lab notebook. If you are here because you are interested in implementing educatETH E.coli in your lab, then our System Implementation and the System Parts pages may be of help to you!
Introduction
For all our cloning procedures we used standard protocols according to SAMBROOK and RUSSELL Molecular Cloning: A Laboratory Manual.
Strains
We used the following E. coli strains:
[http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29|E. coli Top10 (Invitrogen):]
- This strain has a streptomycin resistance
- Genotype: F’ {tetR}, mcrA, Δ(mrr-hsdRMS-mcrBC), φ80 lacZ ΔM15, ΔlacX74, deoR, recA1, araD139 Δ(ara-leu)7679, galU, galK, λ-, rpsL,endA1, nupG
- For further information please [http://openwetware.org/wiki/E._coli_genotypes#TOP10_.28Invitrogen.29| click here]
- References:
- Casdaban, M. and Cohen, S. (1980) J Mol Biol 138:179 PMID 6997493
- Grant, S.G.N. et al. (1990) Proc. Natl. Acad. Sci. USA 87: 4645-4649 PMID 2162051
- Casdaban, M. and Cohen, S. (1980) J Mol Biol 138:179 PMID 6997493
[http://openwetware.org/wiki/E._coli_genotypes#JM101|E. coli JM101:]
- We call them Jimmys
- This strain is the original blue/white cloning strain
- Genotype: glnV44, thi-1, Δ(lac-proAB), F'[lacIqZΔM15 traD36 proAB+]
- For further information please [http://openwetware.org/wiki/E._coli_genotypes#JM101| click here]
- Reference:
- Messing, J. et al. (1981) Nucleic Acids Res. 9, 309; Yanisch-Perron, C., Vieira, J., and Messing, J. (1985) Gene 33, 103
Plasmids
For our system we needed three plasmids with different origins of replication and antibiotic resistances. We decided to take low copy plasmids. We decided to use the following plasmids, which we wanted modify so that they would become compatible to the Biobrick Library multiple cloning site:
Basic plasmids
| Plasmid | Resistances | Copy number | Origin | Map |
|---|---|---|---|---|
| pBR322 | Ampicillin, Tetracyline | 15-20 [1] | pMB1 | |
| pCK01 | Chloramphenicol | 5-12 [1] | pSC101 | |
| pACYC177 | Ampicillin, Kanamycin | 10-12 [1] | p15A |
Changes to the plasmids
In order to get the Biobrick multiple cloning site into the plasmids, we had to make several changes to the plasmids:
| Plasmid | Changes | New name | New resistance | New Map |
|---|---|---|---|---|
| pBR322 |
|
BBa_I739201 |
Ampicillin | |
| pBR322 |
|
BBa_I739203 |
Tetracycline | |
| pCK01 |
|
BBa_I739202 |
Chloramphenicol | |
| pACYC177 |
|
BBa_I739204 |
Kanamycin |
Cloning plan
Parts assignment into plasmids
Three plasmids are used for the EducatETH E.coli system parts as follows:
| plasmid | resistance | copy type | contents | comments |
|---|---|---|---|---|
| pbr322 | ampicillin | high | 1,2,3 | constitutive subsystem |
| pck01 | chloramphenicol | low | 4,5,8,9 | reporting subsystem |
| pacyc177 | kanamycin | low | 6,7,10,11 | learning subsystem, reporting subsystem |
It is important to insert parts responsible for the production of fluorescent proteins in low copy plasmids, as they are potentially harmful for the cell. Unfortunately, working with low copy plasmids makes the procedure more demanding in the lab.
Linkers
Because the plasmids used were not standard plasmids found in the registry, but came from the lab where we work, linkers compatible with the standard BioBrick assembly have to be used in order to work with them. The list of all linkers is the following:
| Linker | Plasmid |
|---|---|
| pbr322-1 | pbr322 |
| pbr322-2 | pbr322 |
| pbr322-3 | pbr322 |
| pbr322-4 | pbr322 |
| pck01 | pck01 |
| pck01-2 | pck01 |
| pacyc177-1 | pacyc177 |
| pacyc177-2 | pacyc177 |
Note that four linkers are tested for pbr322, as two are used for the tetracycline-resistance version of pbr322 and two are used for the ampicillin-resistnace version.
Procedure
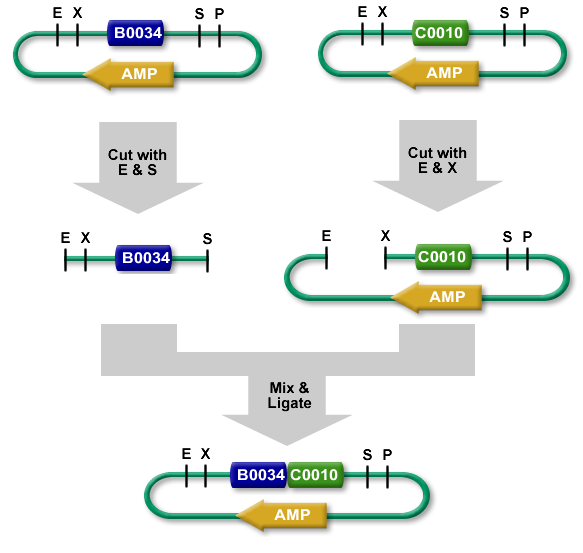
The standard BioBrick assembly will be used to put the parts in the plasmids. Detailed information on how the BioBrick part fabrication works can be found [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication here]. For a shorter explanation of how to assemble 2 parts together check [http://partsregistry.org/Assembly:Standard_assembly here]. Note that the composite part is constructed from the end to the beginning, i.e. each new part is inserted before the existing one. In the following, the plasmid containing the new part to be inserted will be referred to as the donor and the plasmid accepting the new part will be referred to as the acceptor. Composite pars made of parts a and b are denoted a.b.Plasmid 1 (pbr322ap)
- Put parts 1,2,3 in pbr322ap plasmids.
- Merge plasmid containing part 2 (donor) with plasmid containing part 3 (acceptor). You should get a plasmid containing a 2.3 composite part.
- Merge plasmid containing part 1 (donor) with plasmid containing composite part 2.3 (acceptor). You should get a plasmid containing a 1.2.3 composite part.
Plasmid 2 (pck01cm)
- Put parts 4,5,8,9 in pck01cm plasmids.
- Merge plasmid containing part 4 (donor) with plasmid containing part 5 (acceptor). You should get a plasmid containing a 4.5 composite part.
- Merge plasmid containing part 8 (donor) with plasmid containing part 9 (acceptor). You should get a plasmid containing a 8.9 composite part. Note: this step can be done simultaneously with the above.
- Merge plasmid containing composite part 4.5 (donor) with plasmid containing composite part 8.9 (acceptor). You should get a plasmid containing a 4.5.8.9 composite part.
Plasmid 3 (pacyc177km)
- Put parts 6,7,10,11 in pacyc177km plasmids.
- Merge plasmid containing part 6 (donor) with plasmid containing part 7 (acceptor). You should get a plasmid containing a 6.7 composite part.
- Merge plasmid containing part 10 (donor) with plasmid containing part 11 (acceptor). You should get a plasmid containing a 10.11 composite part. Note: this step can be done simultaneously with the above.
- Merge plasmid containing composite part 6.7 (donor) with plasmid containing composite part 10.11 (acceptor). You should get a plasmid containing a 6.7.10.11 composite part.
Labbook
References
[1] [http://www1.qiagen.com/faq/faqview.aspx?faqid=350&SearchText=&FaqCategoryId=0&MenuItemId=0&catalog=1&ProductLineId=1000228 QIAGEN FAQs]
[x] [http://partsregistry.org/Assembly:Standard_assembly Standard Assembly Process]