Edinburgh/DivisionPopper
From 2007.igem.org
(→References) |
|||
| (88 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | '''MENU''' :[[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/References|Background]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design&Implementation]] | [[Edinburgh/DivisionPopper/Modelling|Modelling]] | [[Edinburgh/DivisionPopper/Status|Wet Lab]] | [[Edinburgh/DivisionPopper/SBApproach|Synthetic Biology Approach]] | [[Edinburgh/DivisionPopper/Conclusions|Conclusions]] | |
| - | + | ||
| - | |||
| - | + | The''' Division PoPper''' is a signal generator device that produces an output of [http://partsregistry.org/cgi/htdocs/AbstractionHierarchy/index.cgi PoPS] as a function of bacterial cell division. In simple terms, it is a device that generates a "pulse" of PoPS signal each time a cell undertakes division. Downstream of the device may be a counter device, quantifiable protein production or some other function of choice. The system may for example perform pre-determined actions such as programmed cell death after a set number of cell divisions or being used to calculate the division frequency. | |
| + | The device relies on dif recombinase sites to flip a DNA segment at each cell division. With this project we hope to further analyse cell division and recombinase mechanisms since bacterial cell division is still relatively poorly understood. We plan to construct and ligate the bricks required for a first proof of concept experiments. We model the device using various approaches (deterministic and stochastic modelling) in order to guide lab work and analyse lab data. Modelling is also used to simulate the composition of our device with a counter device. | ||
| + | In presenting our work, we highlight with comments and discussion the application of Synthetic Biology approaches at each work phase. | ||
| - | |||
| - | === | + | {| width="20%" align="right" style="text-align:center" |
| + | |- | ||
| + | |'''Division PoPper device''' | ||
| + | |- | ||
| + | |[[Image:Overview.png|500px]] The Division PoPper device generates a pulse of PoPS signal at each cell division. | ||
| + | |} | ||
| - | + | ===Sections:=== | |
| + | The structure of the wiki for this project is composed of: | ||
| - | This | + | * This [[Edinburgh/DivisionPopper| '''Introduction''']] page has a brief overview. |
| - | + | * The [[Edinburgh/DivisionPopper/Applications|'''Applications''']] page gives some possible uses of the Division PoPper. | |
| + | * The [[Edinburgh/DivisionPopper/References|'''Background''']] page explains the scientific background of our project and lists paper references validating our work. | ||
| + | * The [[Edinburgh/DivisionPopper/Design|'''Design & Implementation''']] page explains the architecture and the mechanisms of the device in terms of abstraction levels and biological processes. | ||
| + | * The [[Edinburgh/DivisionPopper/Modelling|'''Modelling''']] page contains the computational and mathematical investigation of the system behaviour. | ||
| + | * The [[Edinburgh/DivisionPopper/Status|'''Wet Lab''']] page reports our achievements, so far, in the wet lab. | ||
| + | * The section '''[[Edinburgh/DivisionPopper/SBApproach|Synthetic Biology Approach]]''' discusses how we applied the Synthetic Biology approach to the project construction. | ||
| + | * The [[Edinburgh/DivisionPopper/Conclusions|'''Conclusions''']] page summarizes our final consideration of the project. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | <center> | |
| + | ''A menu with the same links is present at the top of each page for facilitating navigation.'' | ||
Latest revision as of 01:38, 27 October 2007
MENU : Introduction | Background | Applications | Design&Implementation | Modelling | Wet Lab | Synthetic Biology Approach | Conclusions
The Division PoPper is a signal generator device that produces an output of [http://partsregistry.org/cgi/htdocs/AbstractionHierarchy/index.cgi PoPS] as a function of bacterial cell division. In simple terms, it is a device that generates a "pulse" of PoPS signal each time a cell undertakes division. Downstream of the device may be a counter device, quantifiable protein production or some other function of choice. The system may for example perform pre-determined actions such as programmed cell death after a set number of cell divisions or being used to calculate the division frequency.
The device relies on dif recombinase sites to flip a DNA segment at each cell division. With this project we hope to further analyse cell division and recombinase mechanisms since bacterial cell division is still relatively poorly understood. We plan to construct and ligate the bricks required for a first proof of concept experiments. We model the device using various approaches (deterministic and stochastic modelling) in order to guide lab work and analyse lab data. Modelling is also used to simulate the composition of our device with a counter device.
In presenting our work, we highlight with comments and discussion the application of Synthetic Biology approaches at each work phase.
| Division PoPper device |
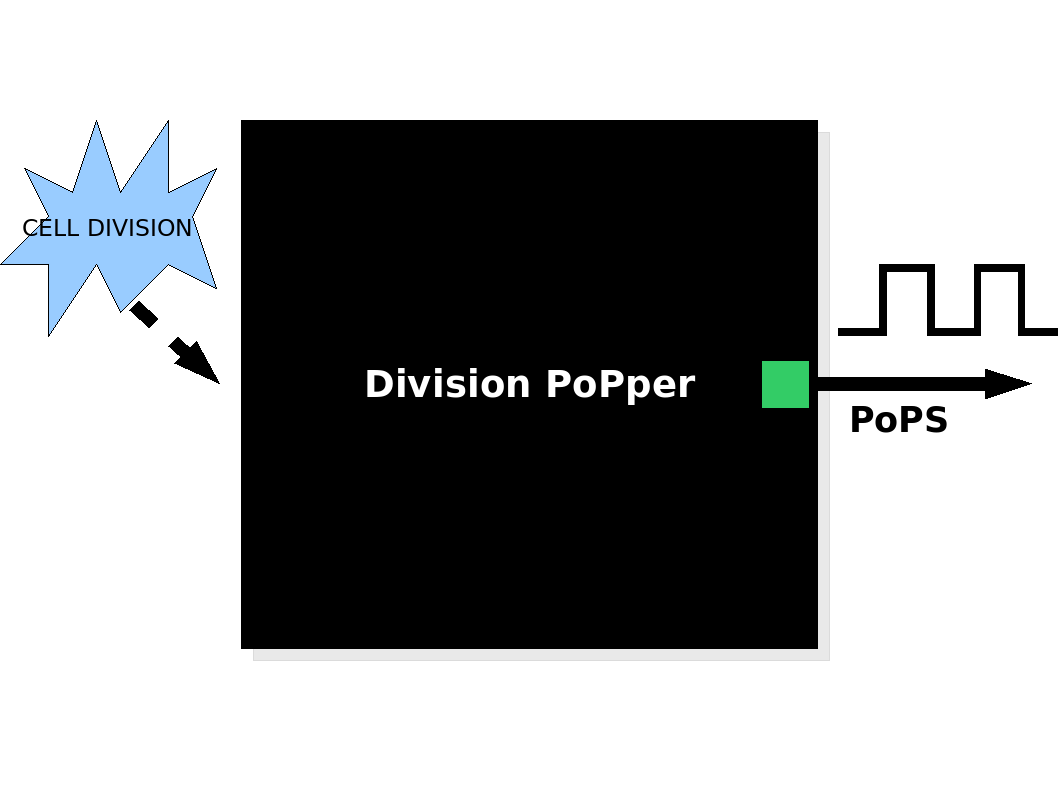 The Division PoPper device generates a pulse of PoPS signal at each cell division. The Division PoPper device generates a pulse of PoPS signal at each cell division.
|
Sections:
The structure of the wiki for this project is composed of:
- This Introduction page has a brief overview.
- The Applications page gives some possible uses of the Division PoPper.
- The Background page explains the scientific background of our project and lists paper references validating our work.
- The Design & Implementation page explains the architecture and the mechanisms of the device in terms of abstraction levels and biological processes.
- The Modelling page contains the computational and mathematical investigation of the system behaviour.
- The Wet Lab page reports our achievements, so far, in the wet lab.
- The section Synthetic Biology Approach discusses how we applied the Synthetic Biology approach to the project construction.
- The Conclusions page summarizes our final consideration of the project.