Edinburgh/DivisionPopper/Conclusions
From 2007.igem.org
| Line 1: | Line 1: | ||
'''MENU''' :[[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/References|Background]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design&Implementation]] | [[Edinburgh/DivisionPopper/Modelling|Modelling]] | [[Edinburgh/DivisionPopper/Status|Wet Lab]] | [[Edinburgh/DivisionPopper/SBApproach|Synthetic Biology Approach]] | [[Edinburgh/DivisionPopper/Conclusions|Conclusions]] | '''MENU''' :[[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/References|Background]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design&Implementation]] | [[Edinburgh/DivisionPopper/Modelling|Modelling]] | [[Edinburgh/DivisionPopper/Status|Wet Lab]] | [[Edinburgh/DivisionPopper/SBApproach|Synthetic Biology Approach]] | [[Edinburgh/DivisionPopper/Conclusions|Conclusions]] | ||
| + | |||
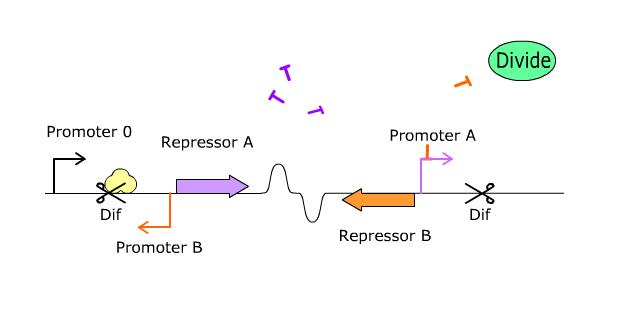
We have designed a switch that works as a function of bacterial division. This switch is naturally oscillatory, as opposed to other engineered switches. Switch frequency depends on what strain you place the device in. The ''dif'' sites are recombinatorial solutions that are employed universally by bacteria, so the PoPper should function well outwith the ''E. coli'' scope. | We have designed a switch that works as a function of bacterial division. This switch is naturally oscillatory, as opposed to other engineered switches. Switch frequency depends on what strain you place the device in. The ''dif'' sites are recombinatorial solutions that are employed universally by bacteria, so the PoPper should function well outwith the ''E. coli'' scope. | ||
| Line 9: | Line 10: | ||
This wiki will be updated until the very moment it freezes, so please watch this space. We keep our fingers crossed. | This wiki will be updated until the very moment it freezes, so please watch this space. We keep our fingers crossed. | ||
| + | |||
| + | |||
| + | |||
| + | [[Image:Popper.JPG]] | ||
| + | |||
| + | Please take some time to visit [http://www.macteria.co.uk/popper.php This page ] for an interactive Flash animation of the final construct! | ||
| + | |||
| + | We look forward to meeting you all at the Jamboree. | ||
Revision as of 03:45, 27 October 2007
MENU : Introduction | Background | Applications | Design&Implementation | Modelling | Wet Lab | Synthetic Biology Approach | Conclusions
We have designed a switch that works as a function of bacterial division. This switch is naturally oscillatory, as opposed to other engineered switches. Switch frequency depends on what strain you place the device in. The dif sites are recombinatorial solutions that are employed universally by bacteria, so the PoPper should function well outwith the E. coli scope.
The Divison PoPper has been a collected team effort. At the time of writing, we're still waiting for the Division Popper to arrive from GeneArt. Delivery has now been delayed by over a month and it looks like final piece in this jigsaw puzzle will be left out for now. The project does not end here, but will be continued as a final year dissertation project by a student, but we would have liked to present results.
All the research we did into the topic still allows us to predict the system outcome. The models (please fill me in on this, Luca)
This wiki will be updated until the very moment it freezes, so please watch this space. We keep our fingers crossed.
Please take some time to visit [http://www.macteria.co.uk/popper.php This page ] for an interactive Flash animation of the final construct!
We look forward to meeting you all at the Jamboree.