ETHZ/Biology
From 2007.igem.org
Stefan Luzi (Talk | contribs) |
(→Current Cloning Status (26.10.07)) |
||
| (196 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:ETHZ_banner.png|830px]] | |
| + | <!-- | ||
| + | <center>[[ETHZ | Main Page]] [[ETHZ/Model | System Modeling]] [[ETHZ/Simulation | Simulations]] [[ETHZ/Biology | System Implementation]] [[ETHZ/Biology/Lab| Lab Notes]] [[ETHZ/Meet_the_team | Meet the Team]] [[ETHZ/Internal | Team Notes]] [[ETHZ/Pictures | Pictures!]]</center><br> | ||
| + | --> | ||
| + | __NOTOC__ | ||
| + | <html> | ||
| + | <script type="text/javascript" src="http://christos.bergeles.net/eth_dropdowntabs.js"> | ||
| - | + | /*********************************************** | |
| + | * Drop Down Tabs Menu- (c) Dynamic Drive DHTML code library (www.dynamicdrive.com) | ||
| + | * This notice MUST stay intact for legal use | ||
| + | * Visit Dynamic Drive at http://www.dynamicdrive.com/ for full source code | ||
| + | ***********************************************/ | ||
| + | </script> | ||
| + | |||
| + | <!-- CSS for Drop Down Tabs Menu #1 --> | ||
| + | <link rel="stylesheet" type="text/css" href="http://christos.bergeles.net/eth_ddcolortabs.css" /> | ||
| + | <div id="colortab" class="ddcolortabs"> | ||
| + | <ul> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ" title="Home" rel="dropmenu_home"><span>Home</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model" title="Modeling" rel="dropmenu_modeling"><span>System Modeling</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation" title="Simulations" rel="dropmenu_simulation"><span>Simulations</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology" title="System Implementation" rel="dropmenu_biology"><span>System Implementation</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team" title="Meet the team" rel="dropmenu_meettheteam"><span>Meet the team</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Pictures" title="Pictures!" rel="dropmenu_pictures"><span>Pictures!</span></a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div class="ddcolortabsline"> </div> | ||
| + | |||
| + | |||
| + | <!--1st drop down menu --> | ||
| + | <div id="dropmenu_home" class="dropmenudiv_a"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Team_Members">Team Members</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Acknowledgments">Acknowledgments</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Site_Map">Site map</a> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <!--2nd drop down menu --> | ||
| + | <div id="dropmenu_modeling" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Model_Overview">Model Overview</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Detailed_Model">Detailed Model</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Final_Model">Final Model</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Modeling_Basics">Modeling Basics Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Mathematical_Model">Mathematical Model</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FSM">FSM View Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FlipFlop">Flip-Flop View Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Parameters">Parameters Page</a> | ||
| + | </div> | ||
| + | |||
| + | <!--3rd drop down menu --> | ||
| + | <div id="dropmenu_simulation" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Simulation_of_Test_Cases">Test Cases</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Sensitivity_Analysis">Sensitivity Analysis</a> | ||
| + | </div> | ||
| + | |||
| + | <!--4th drop down menu --> | ||
| + | <div id="dropmenu_biology" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#The_Complete_System">The Complete System</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#System_Phases">System Phases</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#Current_Cloning_Status">Current Cloning Status</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/parts">System Parts Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/Lab">Lab Notes Page</a> | ||
| + | </div> | ||
| + | |||
| + | <!--5th drop down menu --> | ||
| + | <div id="dropmenu_meettheteam" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#The_ETH_Zurich_07_Team">The ETH Zurich 07 Team</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#Team_Description">Team Description</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Internal">Brainstorming Page</a> | ||
| + | </div> | ||
| + | |||
| + | <script type="text/javascript"> | ||
| + | //SYNTAX: tabdropdown.init("menu_id", [integer OR "auto"]) | ||
| + | tabdropdown.init("colortab", 3) | ||
| + | </script> | ||
| + | |||
| + | </html> | ||
__NOTOC__ | __NOTOC__ | ||
| - | + | = Introduction = | |
| - | + | On this page, you can find an analysis of the function of our system, its biological design, and a list of the parts that make up the system. Under [https://2007.igem.org/ETHZ/Biology/Lab Lab Notes], you can find the ingredients and equipment we used, the electronic version of our lab notebook and a presentation of all the difficulties that we encountered. | |
| - | < | + | educatETH <i>E.coli</i> is a system which can distinguish between [http://openwetware.org/wiki/ATc anhydrotetracycline (aTc)] and [http://openwetware.org/wiki/IPTG Isopropyl-beta-D-thiogalactopyranoside (IPTG)] based on a previous learning phase conducted with the same chemicals and the help of [http://partsregistry.org/Acyl-HSLs acylhomoserine lactone (AHL)]. It is composed of three subsystems: the subsystem of constitutively produced proteins, the learning subsystem and the reporting subsystem. The constitutively produced proteins (LacI, TetR and LuxR) control the learning subsystem. At the core of the latter there exists an extended version of the original toggle switch found in [1]. That is, a multi-inducible toggle switch. The main difference is reflected in the use of double promoters, so that the toggle switch only changes its state when both, one of the two chemicals (aTc/IPTG), and AHL are present. As AHL is only present during the learning phase, the toggle maintains its state during testing/recognition, and thus can “memorize”. AHL can therefore be seen as a training- or learning substance. In the reporting subsystem, four reporters ([http://partsregistry.org/Featured_Parts:Fluorescent_proteins fluorescent proteins]) allow supervision of (1.) the chemical the system was trained with and (2.) if the system recognizes the chemical it is being exposed to in the recognition phase as one it has been previously trained with or not. |
| - | ==== | + | == The Complete System == |
| - | + | ||
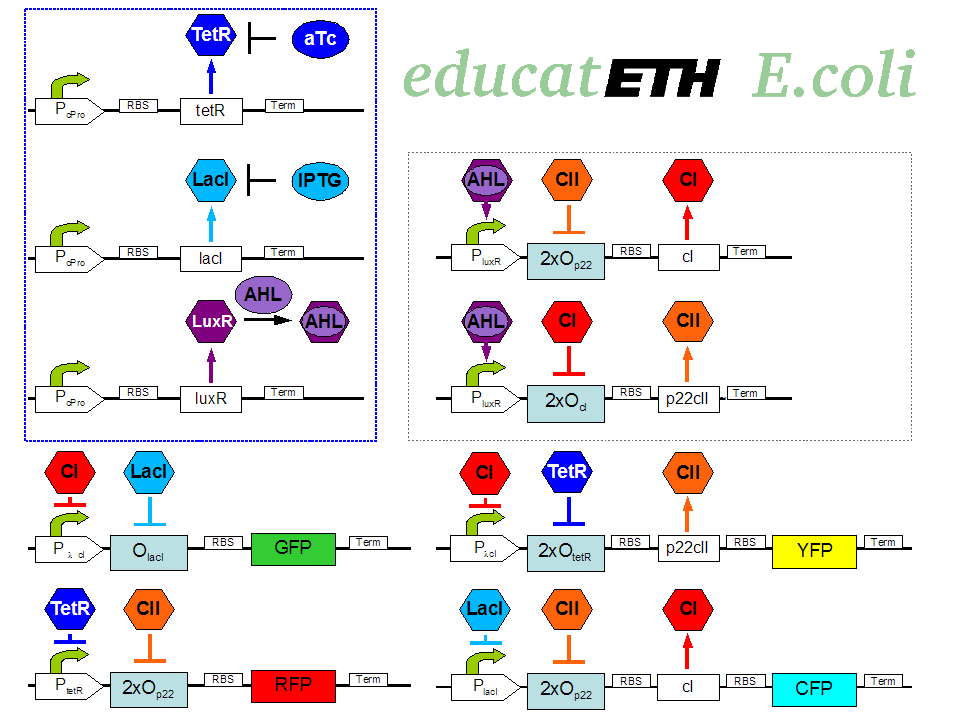
| - | + | <p>[[Image:Biol_system_stand24.10.png|thumb|left|350px|'''Fig. 1:''' Gene interaction network of educatETH ''E.coli'' ]] The biological design of educatETH <i>E.coli</i> is presented in Fig. 1 and below, we clarify the function of all depicted components. (Are you interested in how the complex system of Fig. 1 was modeled? Then visit the [[ETHZ/Model| System Modeling]]!)</p> | |
| - | + | ==== Constitutive Subsystem ==== | |
| - | + | <p>The constitutively produced proteins of the system are LacI, TetR and LuxR. The LuxR part has a special function: when AHL is present, it forms a LuxR-AHL complex which acts on the learning subsystem (more on this later). For now, we will consider that AHL is absent and therefore LuxR cannot activate transcription. The TetR and LacI parts behave similarly: more specifically, the TetR protein in the absence of aTc inhibits the production of p22cII and LacI in the absence of IPTG inhibits the production of cI. When aTc is present, however, the p22cII production is no longer inhibited (and thus p22cII is produced). Correspondingly, cI is produced when IPTG is present.</p> | |
| - | < | + | |
| - | < | + | ==== Learning Subsystem ==== |
| + | |||
| + | <p>The learning subsystem is a toggle switch with two operator sites. The upper part of the toggle (cI production) has operator sites for the LuxR-AHL complex and p22cII (whose production has in turn been induced by aTc). The LuxR-AHL complex induces cI production, whereas p22cII inhibits it. The lower part of the toggle (p22cII production) has operator sites for the LuxR-AHL complex and cI (which has been induced by IPTG). In analogy to the upper part, the LuxR-AHL complex induces production of p22cII and cI inhibits it. Therefore, the switch always requires the presence of the LuxR-AHL complex in order for it to operate. Its state depends on the presence of p22cII and cI in the system, which in turn was caused through the exposure of the system to aTc and IPTG.</p> | ||
| + | |||
| + | ==== Reporting Subsystem ==== | ||
| + | |||
| + | <p>There are four reporters in the system. CFP (more precisely: enhanced CFP, that is ECFP) and YFP (more precisely: enhanced YFP, that is EYFP) are active during the learning phase of the system and show which chemical the system is exposed to during learning, whereas all four reporters (the latter and GFP and RFP) are active during the recognition phase and show if the system is exposed to the same chemical as in learning or not. | ||
| + | More specifically, the YFP production is regulated with help of two operator sites controlled by cI and aTc (TetR inhibitor). cI inhibits the YFP production and aTc induces it. Therefore, YFP is synthesized when the system is exposed to only aTc and cI is not produced within the system (i.e. the system has not been previously exposed to IPTG). The production of the other fluorescent proteins is regulated in a similar manner. Overall, the production of the fluorescent proteins is regulated as follows: | ||
| + | *YFP gets produced when the system is exposed to only aTc and no cI is produced (i.e. the system has ''not'' been previously exposed to IPTG). | ||
| + | *CFP gets produced when the system is exposed to only IPTG and no p22cII is produced (i.e. the system has ''not'' been previously exposed to aTc). | ||
| + | *GFP gets produced when the system is exposed to only IPTG and no cI is produced (i.e. the system has ''not'' been previously exposed to IPTG). | ||
| + | *RFP gets produced when the system is exposed to only aTc and no p22cII is produced (i.e. the system has ''not'' been previously exposed to aTc).</p> | ||
| - | + | This behaviour is visualized in Fig. 2. | |
| - | + | [[Image:ETHzFlowdiagram2.png|center|thumb|350px|<b>Fig. 2</b>: Flow diagram. This figure shows the protocol with which the final system should be tested, as well as the test results in the form of the reported colors. There are three phases the system has to go through: (1) a training or learning phase in which the system learns an input and stores it in its memory, (2) a memory phase in which the system has to keep the content of its memory and, (3) a recognition phase where the output of the system depends on the content of its memory as well as on the current input. |500px]] | |
| - | + | ||
| - | == | + | == System Phases == |
| - | <p> | + | <p>The system operation is divided into three main phases: a learning phase, a memory phase and a recognition phase. During the learning phase, the system is first exposed to one of the two chemicals it is designed to detect (aTc or IPTG). During the memory phase, the specific chemical (aTc or IPTG) is removed and AHL is added to activate the systems internal toggle switch. This maintains the toggle switch to its acquired steady state, which is reported with YFP (if aTc was detected) or CFP (if IPTG was detected). During the recognition phase, the system is exposed to any of the two chemicals (aTc or IPTG), with AHL present. Lets compare the systems toggle switch state with the effect of the newly introduced chemical: the system shows a different response if it has previously been exposed to this certain chemical and reports with the same XFP as in the learning phase (YFP for aTc, CFP for IPTG) or if it recognizes a different chemical and reports with a different XFP (GFP for trained with aTc and recognizing IPTG, RFP for trained with IPTG and recognizing aTc). The following table represents all possible paths that may be taken by the system during all phases of operation according to external stimuli: </p> |
| - | + | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:center; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | |
| - | # | + | |+ '''System phases''' |
| - | + | ! | |
| - | + | !width="44" style="background:#446084; color:white"| aTc | |
| - | + | !width="44" style="background:#446084; color:white"| IPTG | |
| + | !width="44" style="background:#446084; color:white"| AHL | ||
| + | !width="44" style="background:#446084; color:white"| p22cII | ||
| + | !width="44" style="background:#446084; color:white"| cI | ||
| + | ! style="background:#446084; color:white"| Reporting | ||
| + | |- | ||
| + | |colspan="7" style="background:#96c9cf;" align="center"|'''Start''' | ||
| + | |- | ||
| + | | no input | ||
| + | | no | ||
| + | | no | ||
| + | | no | ||
| + | | no | ||
| + | | no | ||
| + | | non | ||
| + | |- | ||
| + | | colspan="7" style="background:#96c9cf;" align="center"| '''Learning''' | ||
| + | |- | ||
| + | | Trained with aTc | ||
| + | | yes | ||
| + | | no | ||
| + | | no | ||
| + | | yes | ||
| + | | no | ||
| + | | YFP | ||
| + | |- | ||
| + | | Trained with IPTG | ||
| + | | no | ||
| + | | yes | ||
| + | | no | ||
| + | | no | ||
| + | | yes | ||
| + | | CFP | ||
| + | |- | ||
| + | | colspan="7" style="background:#96c9cf;" align="center"| '''Memorizing''' | ||
| + | |- | ||
| + | | Trained with aTc | ||
| + | | yes | ||
| + | | no | ||
| + | | yes | ||
| + | | yes | ||
| + | | no | ||
| + | | YFP (fading)<br>finally no color | ||
| + | |- | ||
| + | | Trained with IPTG | ||
| + | | no | ||
| + | | yes | ||
| + | | yes | ||
| + | | no | ||
| + | | yes | ||
| + | | CFP (fading)<br>finally no color | ||
| + | |- | ||
| + | | colspan="7" style="background:#96c9cf;" align="center"| '''Recognition''' | ||
| + | |- | ||
| + | | Trained with aTc<br>Tested with aTc | ||
| + | | yes | ||
| + | | no | ||
| + | | yes | ||
| + | | yes | ||
| + | | no | ||
| + | | YFP | ||
| + | |- | ||
| + | | Trained with aTc<br>Tested with IPTG | ||
| + | | no | ||
| + | | yes | ||
| + | | yes | ||
| + | | yes | ||
| + | | no | ||
| + | | GFP | ||
| + | |- | ||
| + | | Trained with IPTG<br>Tested with IPTG | ||
| + | | no | ||
| + | | yes | ||
| + | | yes | ||
| + | | no | ||
| + | | yes | ||
| + | | CFP | ||
| + | |- | ||
| + | | Trained with IPTG<br>Tested with aTc | ||
| + | | yes | ||
| + | | no | ||
| + | | yes | ||
| + | | no | ||
| + | | yes | ||
| + | | RFP | ||
| + | |- | ||
| + | |||
| + | |} | ||
| - | + | == System Parts == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <p>educatETH <i>E.coli</i> was implemented with 11 basic parts designed by the ETH Zurich team. [https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/parts The list of all the parts, plasmids and strains used] is available. Because the part information is retrieved from the Registry, the page needs some time to load. <br>(Are you interested in this information because you want to implement educatETH <i>E.coli</i> in your lab? Then visit our [https://2007.igem.org/ETHZ/Biology/Lab In the Lab] page!)</p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | == Current Cloning Status (26.10.07) == | |
| - | + | We have sent 9 new parts and 3 new plasmids to the registry. As soon as the missing parts are available in their destined plasmids, we will forward them to the parts registry as well. Additionally, we will be able to test parts of our system next week, using the FACS machine provided by [http://www.facs.ethz.ch Alfredo Franco-Obregón's lab]. | |
| - | + | ||
| - | == | + | == References == |
| - | + | ||
| - | + | [http://www.nature.com/nature/journal/v403/n6767/abs/403339a0.html [1] Gardner TS, Cantor CR and Collins JJ] <i>"Construction of a genetic toggle switch in Escherichia coli"</i>, Nature 403:339–342, 2000<br /> | |
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 23:14, 26 October 2007
Introduction
On this page, you can find an analysis of the function of our system, its biological design, and a list of the parts that make up the system. Under Lab Notes, you can find the ingredients and equipment we used, the electronic version of our lab notebook and a presentation of all the difficulties that we encountered.
educatETH E.coli is a system which can distinguish between [http://openwetware.org/wiki/ATc anhydrotetracycline (aTc)] and [http://openwetware.org/wiki/IPTG Isopropyl-beta-D-thiogalactopyranoside (IPTG)] based on a previous learning phase conducted with the same chemicals and the help of [http://partsregistry.org/Acyl-HSLs acylhomoserine lactone (AHL)]. It is composed of three subsystems: the subsystem of constitutively produced proteins, the learning subsystem and the reporting subsystem. The constitutively produced proteins (LacI, TetR and LuxR) control the learning subsystem. At the core of the latter there exists an extended version of the original toggle switch found in [1]. That is, a multi-inducible toggle switch. The main difference is reflected in the use of double promoters, so that the toggle switch only changes its state when both, one of the two chemicals (aTc/IPTG), and AHL are present. As AHL is only present during the learning phase, the toggle maintains its state during testing/recognition, and thus can “memorize”. AHL can therefore be seen as a training- or learning substance. In the reporting subsystem, four reporters ([http://partsregistry.org/Featured_Parts:Fluorescent_proteins fluorescent proteins]) allow supervision of (1.) the chemical the system was trained with and (2.) if the system recognizes the chemical it is being exposed to in the recognition phase as one it has been previously trained with or not.
The Complete System
The biological design of educatETH E.coli is presented in Fig. 1 and below, we clarify the function of all depicted components. (Are you interested in how the complex system of Fig. 1 was modeled? Then visit the System Modeling!)Constitutive Subsystem
The constitutively produced proteins of the system are LacI, TetR and LuxR. The LuxR part has a special function: when AHL is present, it forms a LuxR-AHL complex which acts on the learning subsystem (more on this later). For now, we will consider that AHL is absent and therefore LuxR cannot activate transcription. The TetR and LacI parts behave similarly: more specifically, the TetR protein in the absence of aTc inhibits the production of p22cII and LacI in the absence of IPTG inhibits the production of cI. When aTc is present, however, the p22cII production is no longer inhibited (and thus p22cII is produced). Correspondingly, cI is produced when IPTG is present.
Learning Subsystem
The learning subsystem is a toggle switch with two operator sites. The upper part of the toggle (cI production) has operator sites for the LuxR-AHL complex and p22cII (whose production has in turn been induced by aTc). The LuxR-AHL complex induces cI production, whereas p22cII inhibits it. The lower part of the toggle (p22cII production) has operator sites for the LuxR-AHL complex and cI (which has been induced by IPTG). In analogy to the upper part, the LuxR-AHL complex induces production of p22cII and cI inhibits it. Therefore, the switch always requires the presence of the LuxR-AHL complex in order for it to operate. Its state depends on the presence of p22cII and cI in the system, which in turn was caused through the exposure of the system to aTc and IPTG.
Reporting Subsystem
There are four reporters in the system. CFP (more precisely: enhanced CFP, that is ECFP) and YFP (more precisely: enhanced YFP, that is EYFP) are active during the learning phase of the system and show which chemical the system is exposed to during learning, whereas all four reporters (the latter and GFP and RFP) are active during the recognition phase and show if the system is exposed to the same chemical as in learning or not. More specifically, the YFP production is regulated with help of two operator sites controlled by cI and aTc (TetR inhibitor). cI inhibits the YFP production and aTc induces it. Therefore, YFP is synthesized when the system is exposed to only aTc and cI is not produced within the system (i.e. the system has not been previously exposed to IPTG). The production of the other fluorescent proteins is regulated in a similar manner. Overall, the production of the fluorescent proteins is regulated as follows:
- YFP gets produced when the system is exposed to only aTc and no cI is produced (i.e. the system has not been previously exposed to IPTG).
- CFP gets produced when the system is exposed to only IPTG and no p22cII is produced (i.e. the system has not been previously exposed to aTc).
- GFP gets produced when the system is exposed to only IPTG and no cI is produced (i.e. the system has not been previously exposed to IPTG).
- RFP gets produced when the system is exposed to only aTc and no p22cII is produced (i.e. the system has not been previously exposed to aTc).
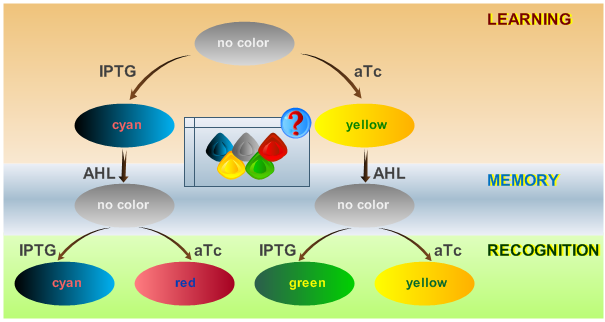
System Phases
The system operation is divided into three main phases: a learning phase, a memory phase and a recognition phase. During the learning phase, the system is first exposed to one of the two chemicals it is designed to detect (aTc or IPTG). During the memory phase, the specific chemical (aTc or IPTG) is removed and AHL is added to activate the systems internal toggle switch. This maintains the toggle switch to its acquired steady state, which is reported with YFP (if aTc was detected) or CFP (if IPTG was detected). During the recognition phase, the system is exposed to any of the two chemicals (aTc or IPTG), with AHL present. Lets compare the systems toggle switch state with the effect of the newly introduced chemical: the system shows a different response if it has previously been exposed to this certain chemical and reports with the same XFP as in the learning phase (YFP for aTc, CFP for IPTG) or if it recognizes a different chemical and reports with a different XFP (GFP for trained with aTc and recognizing IPTG, RFP for trained with IPTG and recognizing aTc). The following table represents all possible paths that may be taken by the system during all phases of operation according to external stimuli:
| aTc | IPTG | AHL | p22cII | cI | Reporting | |
|---|---|---|---|---|---|---|
| Start | ||||||
| no input | no | no | no | no | no | non |
| Learning | ||||||
| Trained with aTc | yes | no | no | yes | no | YFP |
| Trained with IPTG | no | yes | no | no | yes | CFP |
| Memorizing | ||||||
| Trained with aTc | yes | no | yes | yes | no | YFP (fading) finally no color |
| Trained with IPTG | no | yes | yes | no | yes | CFP (fading) finally no color |
| Recognition | ||||||
| Trained with aTc Tested with aTc | yes | no | yes | yes | no | YFP |
| Trained with aTc Tested with IPTG | no | yes | yes | yes | no | GFP |
| Trained with IPTG Tested with IPTG | no | yes | yes | no | yes | CFP |
| Trained with IPTG Tested with aTc | yes | no | yes | no | yes | RFP |
System Parts
educatETH E.coli was implemented with 11 basic parts designed by the ETH Zurich team. The list of all the parts, plasmids and strains used is available. Because the part information is retrieved from the Registry, the page needs some time to load.
(Are you interested in this information because you want to implement educatETH E.coli in your lab? Then visit our In the Lab page!)
Current Cloning Status (26.10.07)
We have sent 9 new parts and 3 new plasmids to the registry. As soon as the missing parts are available in their destined plasmids, we will forward them to the parts registry as well. Additionally, we will be able to test parts of our system next week, using the FACS machine provided by [http://www.facs.ethz.ch Alfredo Franco-Obregón's lab].
References
[http://www.nature.com/nature/journal/v403/n6767/abs/403339a0.html [1] Gardner TS, Cantor CR and Collins JJ] "Construction of a genetic toggle switch in Escherichia coli", Nature 403:339–342, 2000