Alberta
From 2007.igem.org
(→'''The Project: Plan B''') |
(→'''The Team''') |
||
| (22 intermediate revisions not shown) | |||
| Line 22: | Line 22: | ||
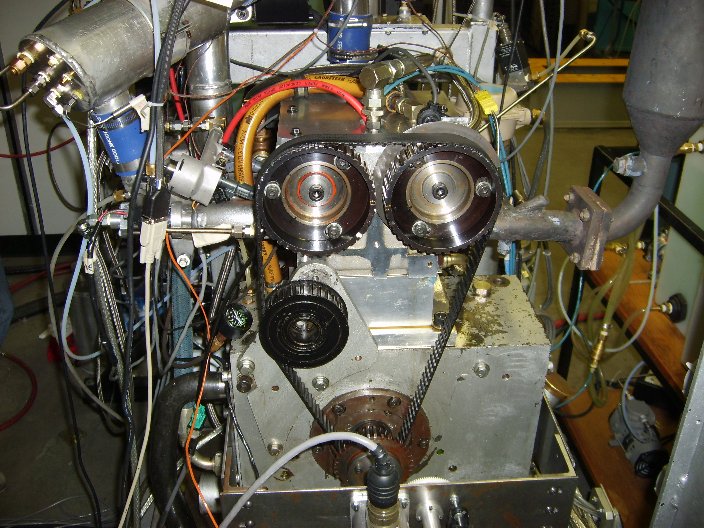
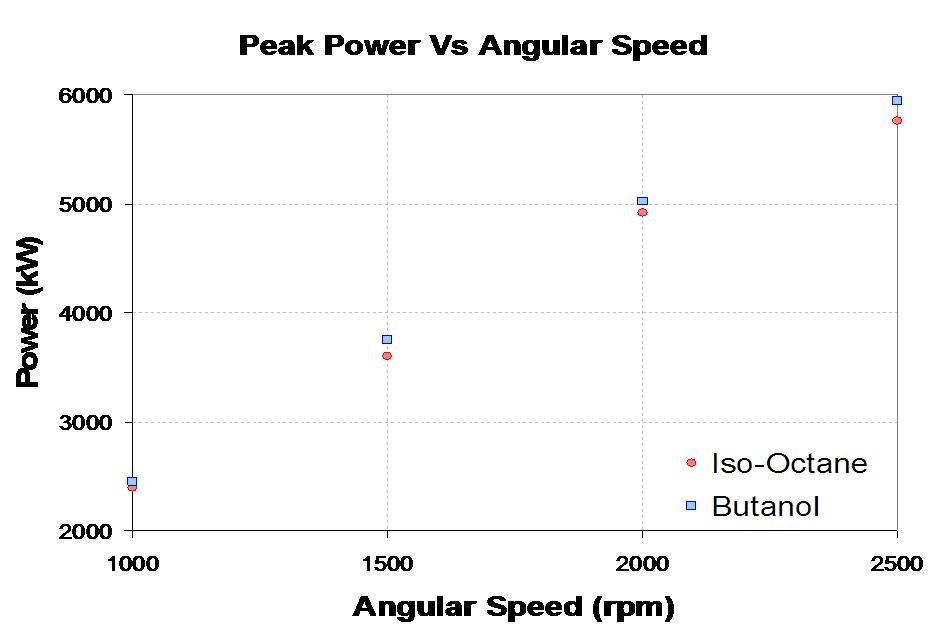
| - | The Butanerds wanted to further evaluate the use of butanol as a fuel in standard spark ignition engines. With the assistance of The University of Alberta's Engine Control Lab in Mechanical Engineering we were able to run a peak power test comparing butanol to iso-octane, a standard test measurement. Each fuel was burned at a | + | The Butanerds wanted to further evaluate the use of butanol as a fuel in standard spark ignition engines. With the assistance of The University of Alberta's Engine Control Lab in Mechanical Engineering we were able to run a peak power test comparing butanol to iso-octane, a standard test measurement. Each fuel was burned at a stoichiometric air to fuel ratio at 4 different angular speeds. The experimental setup and results can be seen below. |
[[image: alberta_engine1.jpeg|thumb|left|275px| Engine Test Setup Front View]] | [[image: alberta_engine1.jpeg|thumb|left|275px| Engine Test Setup Front View]] | ||
| Line 31: | Line 31: | ||
== '''The Project: Plan B''' == | == '''The Project: Plan B''' == | ||
| - | We propose the use of butanol as the leading biofuel for use in internal combustion engines. Specifically, we intend to genetically engineer ''Escherichia coli'' bacteria to convert | + | We propose the use of butanol as the leading biofuel for use in internal combustion engines. Specifically, we intend to genetically engineer ''Escherichia coli'' bacteria to convert biomass into butanol for use as an energy source. This will be accomplished by introducing the genes responsible for butanol production from ''Clostridium acetobutylicum'' (i.e. endogenous butanoate pathway) into ''E. coli''. Furthermore, we hope to increase ''E. coli'''s tolerance to solvents such as butanol. |
| - | + | I: Transforming genes in ''C. acetobutylicum'''s butanoate pathway into ''E.coli''<br> | |
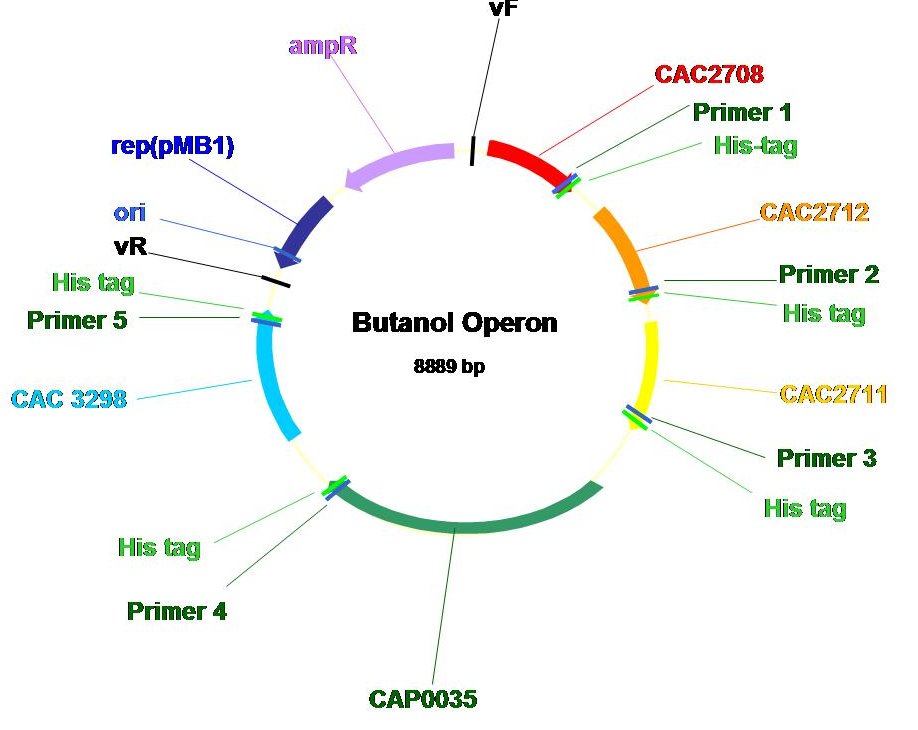
| + | Genes encoding the enzymes in the ''C. acetobutylicum'' butanoate metabolism pathway were identified using the KEGG database (KEGG number-ca00650: http://www.kegg.com/dbget-bin/www_bget?path:cac00650). Our cloning stragegy is to incorporate all the genes in a single operon with respective inducible promoter and ribosome binding site in a plasmid. Such construct enables easy transformation of multiple genes simultaneously into ''E.coli'' and allows the coordinated expression of genes within the operon. <br> | ||
| - | + | After receiving the commercially synthesized coding sequences of the genes of interest, the coding sequences are restricted with proper BioBrick prefixes and suffixes out of the original plasmid and cloned into B0034 (ribosome binding site). Double digest (Xba I and Spe I) as well as automated sequencing reactions are performed to verify the proper insertion of the coding sequences and the presence of a ribosome binding site at the 5' end of each coding sequence. <br> | |
| + | The operon, shown below, consists of all of the genes in the pathway (except ''E. coli'''s thiolase). Note that we purposedly added a Histamine tag to the carboxy terminus of each gene product such that we could use Nickel-NTA column to purify the tagged proteins and subsequently analyze protein expression and enzymatic activity. | ||
| + | [[image: alberta_butanol_Operon.jpeg|thumb|center|400px|The Butanol Operon]] | ||
| - | Concurrently, we are investigating the | + | II: Developing butanol tolerance with the mutagenic and toxic compound--Ethylnitrosourea ENU<br> |
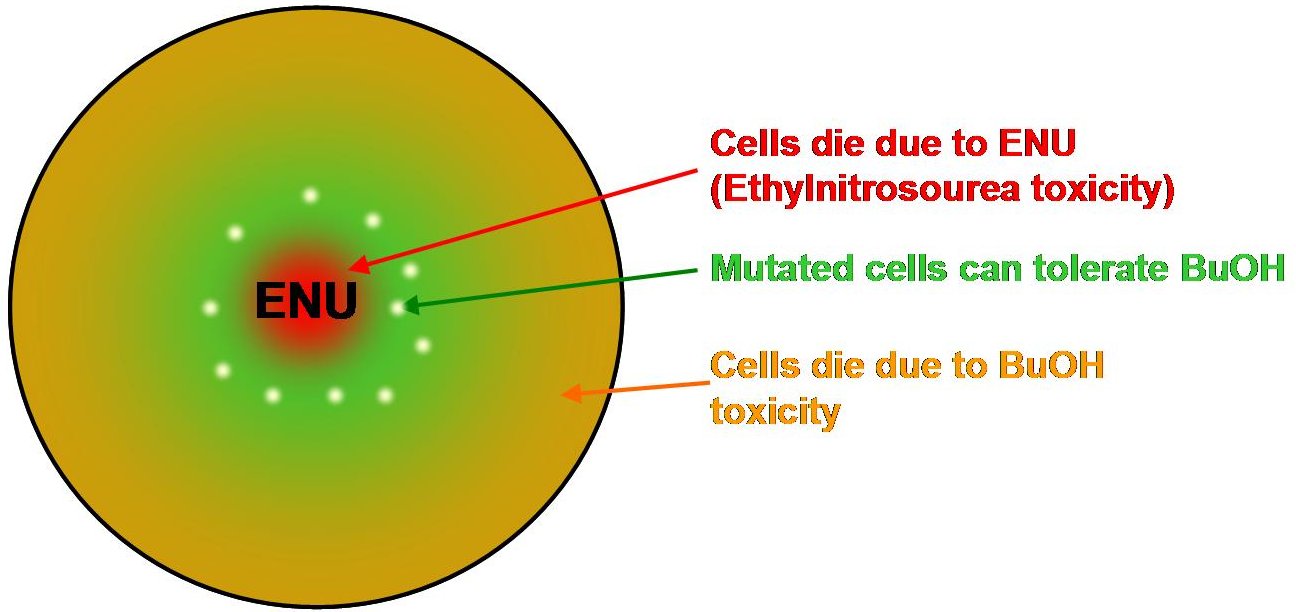
| + | The concept is to make butanol-agar plates of various butanol concentrations and place an ENU disk in the centre. We expect two ring-shaped kill zones. The first where bacteria die due to the toxicity of ENU at the centre of the plate. The second around the outer edge of the butanol-agr plate where bacteria die due to butanol toxicity. In between these two zones we hope to see some cells that would be mutated in a benifical way to render them resistant to butanol. | ||
| + | |||
| + | [[image: alberta_butanolplate2.jpeg|400px|thumb|center|Proposed Butanol-ENU Plate for Mutagenisis of ''E. coli'']] | ||
| + | |||
| + | |||
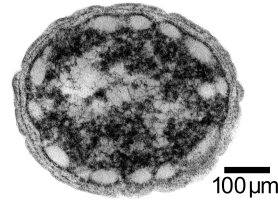
| + | Concurrently, we are investigating the possibiilty of introducing the butanol production pathway into a photoautotrophic bacterium, ''Chlorobium tepidum''. ''C. tepidum'' is a green sulfur bacterium that is strictly anaerobic and uses sulfur compounds as terminal electron donors. ''C. tepidum'' is a moderately thermophillic bacterium, growing at 40 degrees Celsius (104 degrees Fahrenheit), and requires low light conditions for optimal growth. These bacteria grow well in a defined medium, utilizing the reverse/reductive tricarboxylic acid (TCA) cycle to build up carbohydrates from carbon dioxide. In addition, the genome of this bacterium has been completely sequenced. For a more comprehensive overview of ''Chlorobium tepidum'' go to [[http://www.bmb.psu.edu/faculty/bryant/lab/chlorobiumtepidum/index.html ''here'']]. Images below from http://www.bmb.psu.edu/faculty/bryant/lab/chlorobiumtepidum/index.html | ||
[[Image:spacer.jpeg|left]][[Image:Alberta_micrograph.gif|thumb|300px|left|Figure 1:''Chlorobium tepidum'' Transmission Micrograph Image]] | [[Image:spacer.jpeg|left]][[Image:Alberta_micrograph.gif|thumb|300px|left|Figure 1:''Chlorobium tepidum'' Transmission Micrograph Image]] | ||
| Line 44: | Line 53: | ||
<br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br> | ||
| - | |||
| Line 56: | Line 64: | ||
| - | Therefore, to obtain a rough "first-order" approximation for the behavior of the pathway, we considered | + | Therefore, to obtain a rough "first-order" approximation for the behavior of the pathway, we considered three factors: |
1) Stoichiometric factors (both redox and carbon)<br> | 1) Stoichiometric factors (both redox and carbon)<br> | ||
| - | 2) Compared the relative production rates of the various end products of the glucose pathway in ''E. coli'' as found in previous experimentation to the production rates of butanol in ''C. acetobutylicum''. | + | 2) Compared the relative production rates of the various end products of the glucose pathway in ''E. coli'' as found in previous experimentation to the production rates of butanol in ''C. acetobutylicum''. <br> |
3) We have also a fuel E. coli Stoichometric Matrix, and intend to stoichiometrically model the system in order to optimize butanol production once the operon is in the system. | 3) We have also a fuel E. coli Stoichometric Matrix, and intend to stoichiometrically model the system in order to optimize butanol production once the operon is in the system. | ||
| Line 67: | Line 75: | ||
In order to further develop the system, we hope a full metabolic stochiometric model can be eventually realized for this project. This would allow optimization of the system in order to maximize the flux through the butanol pathway. | In order to further develop the system, we hope a full metabolic stochiometric model can be eventually realized for this project. This would allow optimization of the system in order to maximize the flux through the butanol pathway. | ||
| - | |||
== '''Project Timeline''' == | == '''Project Timeline''' == | ||
| Line 76: | Line 83: | ||
'''Legend: ''' <br> | '''Legend: ''' <br> | ||
| - | Benny - <br> | + | Benny - Butyryl CoA dehydrogenase<br> |
| - | Enny - <br> | + | Enny - Enoyl CoA hydratase<br> |
| - | Buddy - <br> | + | Buddy - Butanol dehydrogenase<br> |
| - | Betty - <br> | + | Betty - Beta-hydraoxy butyryl CoA dehydrogenase<br> |
| - | Deisel Blaze - | + | Deisel Blaze - Butyraldehyde dehydrogenase<br> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
== '''Lab Book and Calendar''' == | == '''Lab Book and Calendar''' == | ||
| Line 105: | Line 104: | ||
[[Alberta/Calender/October|October 2007]] | [[Alberta/Calender/October|October 2007]] | ||
| - | ' | + | Meetings (Agenda's, Minutes, Action Items): |
| + | [[Alberta/Calender/Meetings|Meeting Information]] | ||
| + | |||
| + | |||
| + | Online Form: | ||
| + | |||
| + | '''[http://butanerds.myfreeforum.org The Butanerd Online Forum]''' is up and running. Feel free to post and check for posts here. Make sure you register! | ||
== '''The Team''' == | == '''The Team''' == | ||
| Line 114: | Line 119: | ||
Our team has a rich background in biology, biochemistry and engineering. To compliment our diversity we also have advisors who have a wealth of knowledge in research and applications of genetic engineering. For more information about the group, check out the '''[[Alberta/Members|University of Alberta's iGEM Team Members Page]]'''. | Our team has a rich background in biology, biochemistry and engineering. To compliment our diversity we also have advisors who have a wealth of knowledge in research and applications of genetic engineering. For more information about the group, check out the '''[[Alberta/Members|University of Alberta's iGEM Team Members Page]]'''. | ||
| + | |||
| + | [[Advisors:]] <br> | ||
| + | Dr. Michael Deyholos deyholos[at]ualberta.ca <br> | ||
| + | Dr. Douglas Ridgway ridgway[at]ualberta.ca <br> | ||
| + | Dr. Wayne Materi wayne.materi[at]ualberta.ca <br> | ||
| + | James Maclagan james.maclagan[at]ualberta.ca<br> | ||
| + | |||
| + | [[Team:]] <br> | ||
| + | Michelle Chan – Biological Sciences mcchan[at]ualberta.ca <br> | ||
| + | Erin Dul – Biochemistry edul[at]ualberta.ca <br> | ||
| + | Jason Gardiner – Botany jlg6[at]ualberta.ca <br> | ||
| + | Nick Glass – Mechanical Engineering nglass[at]ualberta.ca <br> | ||
| + | Graduate student in Electrical Engineering<br> | ||
| + | Veronica Houston – Mechanical Engineering vhouston[at]ualberta.ca <br> | ||
| + | Nikhil Kumar – Biological Sciences nkumar[at]ualberta.ca <br> | ||
| + | Matt LaFleche – Engineering Physics mjl3[at]ualberta.ca <br> | ||
| + | Alex Lam – Pharmacology pokman[at]ualberta.ca <br> | ||
| + | Justin Pahara – Immunology & Infection jpahara[at]ualberta.ca <br> | ||
| + | Graduate student in Cell Biology <br> | ||
| + | Celine Zeng – Pharmacology youci[at]ualberta.ca <br> | ||
| + | |||
| + | We thank you for your interest, and further inquiries can be directed to igem[at]ualberta.ca <br> | ||
Our University of Alberta Student Group Constitution can be found '''[[alberta/constitution|here]]''' | Our University of Alberta Student Group Constitution can be found '''[[alberta/constitution|here]]''' | ||
| + | <nowiki>Insert non-formatted text here</nowiki> | ||
== '''Edmonton''' == | == '''Edmonton''' == | ||
Latest revision as of 01:33, 10 January 2008

The Offical Wiki of the 2007 University of Alberta iGEM Team
Contents |
Background: Biofuels
With growing concerns of global greenhouse gasses, the global energy market is in a continual push for the development of renewable energy sources. Specifically, two fuels have dominated the media: biodiesel and ethanol. However, both of these fuels have shortcomings in terms of being a viable fuel source.
Biodiesel is a fuel produced from the vegetable oils of crops that can be used in an engine system very similar to a traditional diesel engine. However, vegetable oils in crops only make up a small portion of the biomass of the plants, ultimately producing low yields of fuel per acre of crops. As such, it is more economically sound to use these resources for food production.
There has been huge attention to the use of ethanol in the typical auto cycle engine. In many places it is already being blended with gasoline to create a hybrid fuel source. However, running an engine on pure ethanol is beset by several major obstacles. Firstly, ethanol is miscible with water at any concentration, which creates long term storage corrosion issues. Secondly, ethanol engines must be designed to expect water vapor unlike their gasoline counterparts. Ethanol also has significantly different thermodynamic properties than gasoline such as a lower energy density and different vapor properties, which would reduce the economical advantage of using ethanol as a primary fuels source.
We propose using a different fuel source to eventually replace gasoline, butanol. Butanol is a superior to ethanol as a replacement for petroleum gasoline. With a low vapor pressure, high energy density, and a gasoline-like octane rating, it can be blended into existing gasoline at much higher proportions than ethanol without compromising performance, mileage, organic pollution standards. Blending butanol with gasoline also prevents major modifications of the fuel-air ratio and modifications to the fuel system. Butanol also is immiscible with water at concentrations higher than 7%, alleviating storage concerns as well as offering the possiblity of phase separation which would realize huge cost savings in terms of production.
More information on the summary of biofuels viability and the inspiration to our project can be found here.
The Butanerds wanted to further evaluate the use of butanol as a fuel in standard spark ignition engines. With the assistance of The University of Alberta's Engine Control Lab in Mechanical Engineering we were able to run a peak power test comparing butanol to iso-octane, a standard test measurement. Each fuel was burned at a stoichiometric air to fuel ratio at 4 different angular speeds. The experimental setup and results can be seen below.
The Project: Plan B
We propose the use of butanol as the leading biofuel for use in internal combustion engines. Specifically, we intend to genetically engineer Escherichia coli bacteria to convert biomass into butanol for use as an energy source. This will be accomplished by introducing the genes responsible for butanol production from Clostridium acetobutylicum (i.e. endogenous butanoate pathway) into E. coli. Furthermore, we hope to increase E. coli's tolerance to solvents such as butanol.
I: Transforming genes in C. acetobutylicum's butanoate pathway into E.coli
Genes encoding the enzymes in the C. acetobutylicum butanoate metabolism pathway were identified using the KEGG database (KEGG number-ca00650: http://www.kegg.com/dbget-bin/www_bget?path:cac00650). Our cloning stragegy is to incorporate all the genes in a single operon with respective inducible promoter and ribosome binding site in a plasmid. Such construct enables easy transformation of multiple genes simultaneously into E.coli and allows the coordinated expression of genes within the operon.
After receiving the commercially synthesized coding sequences of the genes of interest, the coding sequences are restricted with proper BioBrick prefixes and suffixes out of the original plasmid and cloned into B0034 (ribosome binding site). Double digest (Xba I and Spe I) as well as automated sequencing reactions are performed to verify the proper insertion of the coding sequences and the presence of a ribosome binding site at the 5' end of each coding sequence.
The operon, shown below, consists of all of the genes in the pathway (except E. coli's thiolase). Note that we purposedly added a Histamine tag to the carboxy terminus of each gene product such that we could use Nickel-NTA column to purify the tagged proteins and subsequently analyze protein expression and enzymatic activity.
II: Developing butanol tolerance with the mutagenic and toxic compound--Ethylnitrosourea ENU
The concept is to make butanol-agar plates of various butanol concentrations and place an ENU disk in the centre. We expect two ring-shaped kill zones. The first where bacteria die due to the toxicity of ENU at the centre of the plate. The second around the outer edge of the butanol-agr plate where bacteria die due to butanol toxicity. In between these two zones we hope to see some cells that would be mutated in a benifical way to render them resistant to butanol.
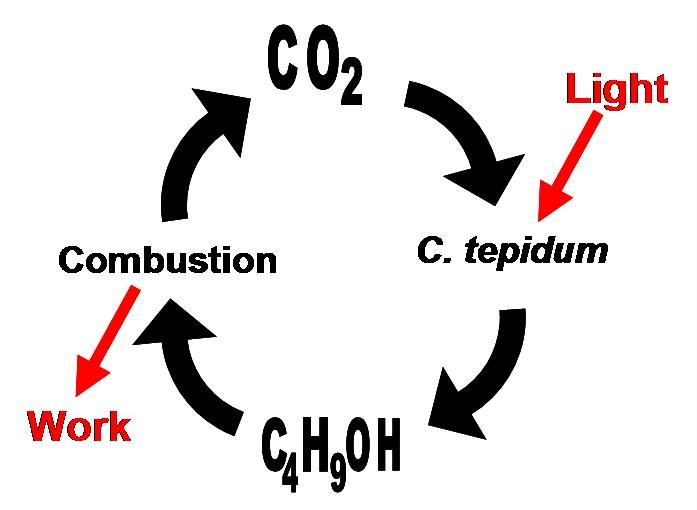
Concurrently, we are investigating the possibiilty of introducing the butanol production pathway into a photoautotrophic bacterium, Chlorobium tepidum. C. tepidum is a green sulfur bacterium that is strictly anaerobic and uses sulfur compounds as terminal electron donors. C. tepidum is a moderately thermophillic bacterium, growing at 40 degrees Celsius (104 degrees Fahrenheit), and requires low light conditions for optimal growth. These bacteria grow well in a defined medium, utilizing the reverse/reductive tricarboxylic acid (TCA) cycle to build up carbohydrates from carbon dioxide. In addition, the genome of this bacterium has been completely sequenced. For a more comprehensive overview of Chlorobium tepidum go to [[http://www.bmb.psu.edu/faculty/bryant/lab/chlorobiumtepidum/index.html here]]. Images below from http://www.bmb.psu.edu/faculty/bryant/lab/chlorobiumtepidum/index.html
The advantage of manipulating this organism for butanol production is a matter of energy input and output. Rather than having to utilize vast amounts of food stocks, such as grains or sugars, a photoautotroph will fix carbon dioxide into the complex carbohydrates required for butanol production. Theoretically, if the only fuel on our planet was butanol, and it was produced in this manner, there would be little net carbon dioxide produced.
Modeling Efforts
Initially our plan to model the efficiency of the butanol production hinged on developing a complete model for the entire glucose pathway in an E. coli cell using Michaelis-Menten kinetics. After further review, this proved to be beyond the scope of our project's timeline due both to the large number of species and reactions present in the system (as well as their non-linear behavior) and the relative difficulty in finding experimental kinetic information for some of the reactions in our pathway.
Therefore, to obtain a rough "first-order" approximation for the behavior of the pathway, we considered three factors:
1) Stoichiometric factors (both redox and carbon)
2) Compared the relative production rates of the various end products of the glucose pathway in E. coli as found in previous experimentation to the production rates of butanol in C. acetobutylicum.
3) We have also a fuel E. coli Stoichometric Matrix, and intend to stoichiometrically model the system in order to optimize butanol production once the operon is in the system.
From these factors we can obtain an indicator of how likely butanol will be produced.
In order to further develop the system, we hope a full metabolic stochiometric model can be eventually realized for this project. This would allow optimization of the system in order to maximize the flux through the butanol pathway.
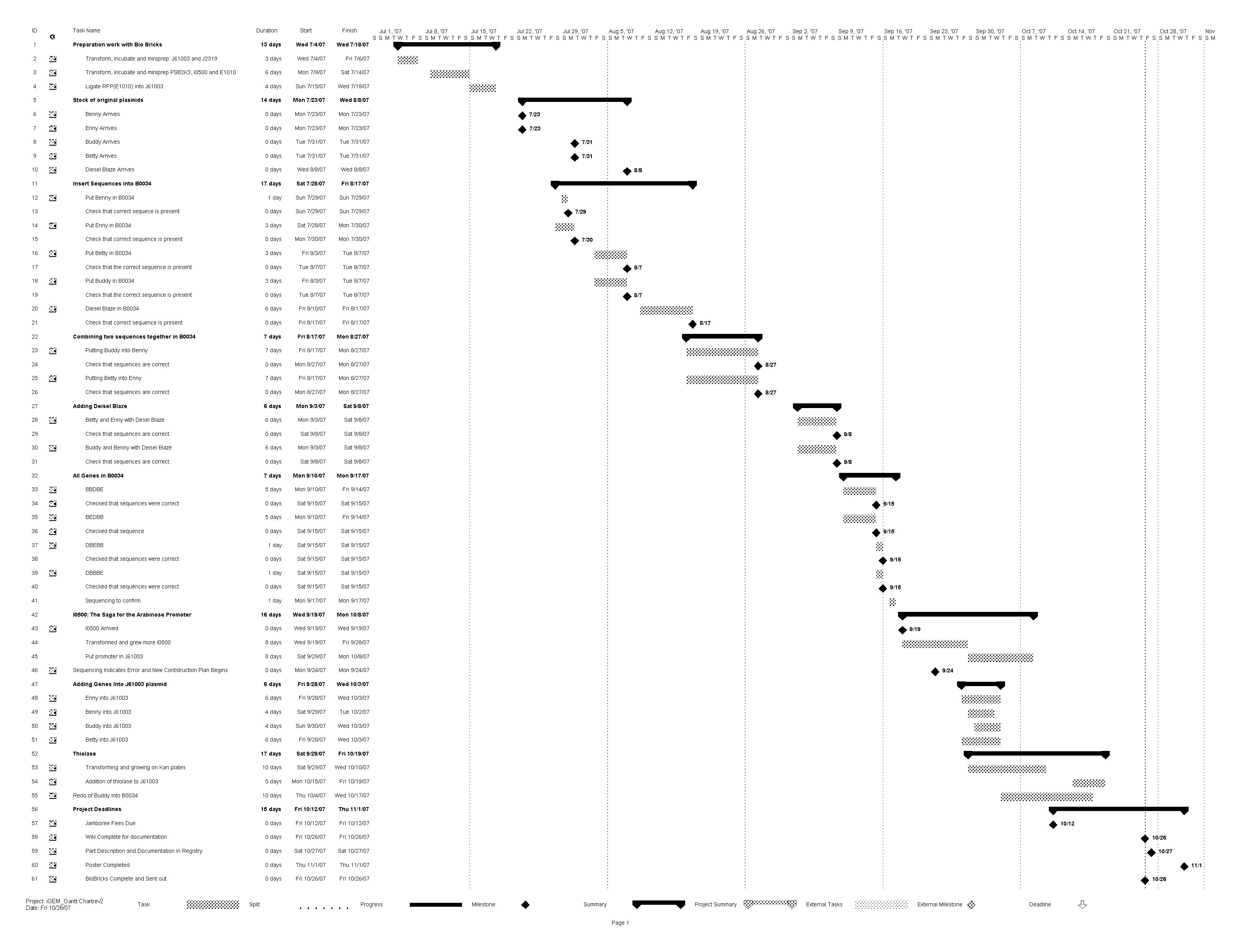
Project Timeline
The GANTT Chart Above details the schedule for the lab work that contributed to Plan B. Beginning in July and finishing with our final work in October, the schedule includes details on our different methods to compose an operon that would meet our objectives. In order to read the GANTT Chart for the Lab work of Plan B, please refer to the legend below.
Legend:
Benny - Butyryl CoA dehydrogenase
Enny - Enoyl CoA hydratase
Buddy - Butanol dehydrogenase
Betty - Beta-hydraoxy butyryl CoA dehydrogenase
Deisel Blaze - Butyraldehyde dehydrogenase
Lab Book and Calendar
Butanerd Event Calendar can be found below:
[http://www.ualberta.ca/~mjl3/UofAIgemProtocols.pdf The Lab Protocols]
Meetings (Agenda's, Minutes, Action Items):
Online Form:
[http://butanerds.myfreeforum.org The Butanerd Online Forum] is up and running. Feel free to post and check for posts here. Make sure you register!
The Team

Our team has a rich background in biology, biochemistry and engineering. To compliment our diversity we also have advisors who have a wealth of knowledge in research and applications of genetic engineering. For more information about the group, check out the University of Alberta's iGEM Team Members Page.
Advisors:
Dr. Michael Deyholos deyholos[at]ualberta.ca
Dr. Douglas Ridgway ridgway[at]ualberta.ca
Dr. Wayne Materi wayne.materi[at]ualberta.ca
James Maclagan james.maclagan[at]ualberta.ca
Team:
Michelle Chan – Biological Sciences mcchan[at]ualberta.ca
Erin Dul – Biochemistry edul[at]ualberta.ca
Jason Gardiner – Botany jlg6[at]ualberta.ca
Nick Glass – Mechanical Engineering nglass[at]ualberta.ca
Graduate student in Electrical Engineering
Veronica Houston – Mechanical Engineering vhouston[at]ualberta.ca
Nikhil Kumar – Biological Sciences nkumar[at]ualberta.ca
Matt LaFleche – Engineering Physics mjl3[at]ualberta.ca
Alex Lam – Pharmacology pokman[at]ualberta.ca
Justin Pahara – Immunology & Infection jpahara[at]ualberta.ca
Graduate student in Cell Biology
Celine Zeng – Pharmacology youci[at]ualberta.ca
We thank you for your interest, and further inquiries can be directed to igem[at]ualberta.ca
Our University of Alberta Student Group Constitution can be found here Insert non-formatted text here
Edmonton
Welcome to Edmonton!

For more on the city of Edmonton click [http://www.ualberta.ca/~mjl3/About.html here].
Sponsors
We would like to thank all of our sponsers for their gracious support and our advisors for invaluble advise.
Major Sponsors

|
Sponsors

|
Advice
Andrew Hessel - Alberta Ingenuity Mentor
Dr. Jonathan Dennis - University of Alberta
Dr. Perrin Beatty - University of Alberta
Dr. David Bressler - University of Alberta
Dr. Charles Lucy - University of Alberta
Dr. Gregory Kiema - University of Alberta
Dr. Mark S. Peppler - University of Alberta
Dr. James Harynuk - University of Alberta
Dr. Federick West-University of Alberta
Dr. Todd Lowary-University of Alberta
Desiree Schell - University of Alberta CJSR
Dr. Jeff Fuller - University of Alberta/Capital Health
Dr. Julia Foght - University of Alberta
Dr. David Checkel - University of Alberta
Adrian Audiet - University of Alberta
Dr. Koch - University of Alberta
Dr. Donald Bryant - Pennsylvania State
Dr. Gaozhong Shen - Pennsylvania State
Amaya Garcia - Pennsylvania State
A list of experimental supplies we need can be found here
External Links
[http://www.albertaingenuity.ca/ Alberta Ingenuity Fund]
[http://www.ualberta.ca The University of Alberta Hompage]
[http://www.mece.ualberta.ca/~ckoch/ Dr. Koch's Engine Lab]
[http://www.systems-biology.org/cd Cell Designer Homepage]
[http://www.biomodels.org Biomodels Home Page]