Caltech/Project/Riboregulator
From 2007.igem.org
(→Riboregulator Design) |
|||
| (One intermediate revision not shown) | |||
| Line 15: | Line 15: | ||
==Riboregulators== | ==Riboregulators== | ||
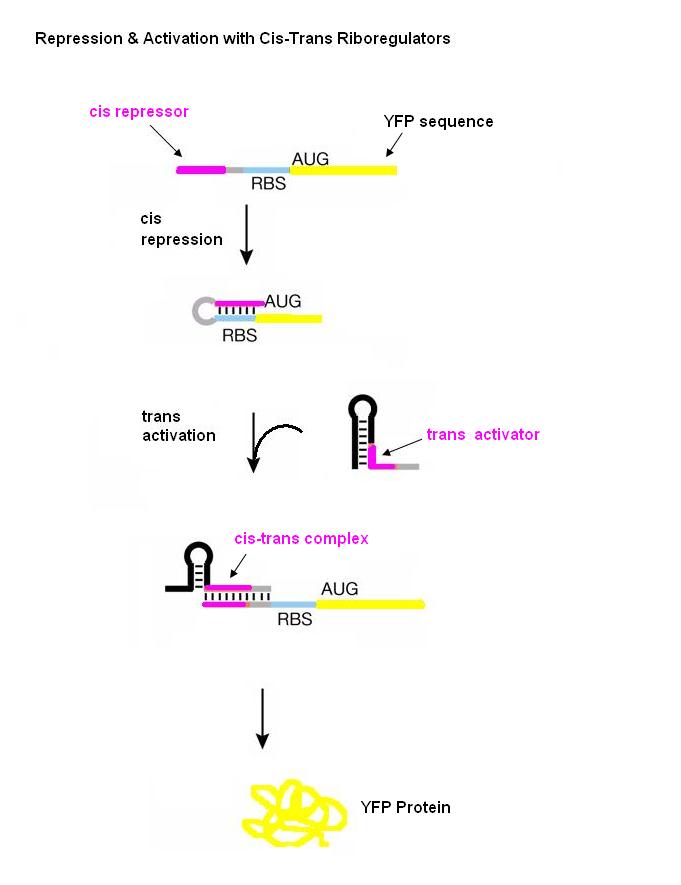
| - | New insights into this regulatory role of RNA have | + | New insights into this regulatory role of RNA have spurred the development of new RNA molecules which control cell behavior through interactions with other RNAs or with small molecule ligands. One such engineering approach which enables post-transcriptional control of gene expression is the riboregulator developed by Collins and coworkers. The riboregulator is composed of two interacting parts, a cis-repressive sequence and a trans-activating sequence. The cis sequence is placed directly upstream of the ribosome binding site (RBS) of the gene regulated by the riboregulator and is complimentary to the RBS. This sequence forms a stem-loop structure in the 5’-untranslated region of the mRNA which prevents ribosome binding and thus translation. The trans RNA is complementary to the cis sequence. When the trans RNA is present, it binds to the cis sequence, the RBS site becomes exposed, and the ribosome can bind and translate the mRNA. |
| - | ==Riboregulator Integration in Our Phage/''E. coli'' System== | + | ===Riboregulator Integration in Our Phage/''E. coli'' System=== |
| - | + | We wish for the riboregulator to determine what happens to a cell once it is infected by our phage. We knock out the native copy of a key developmental gene (N, Q, or cro) and then add it back under the control of the cis-repressor. When the phage infects a wild-type ''E. coli'' cell, nothing should happen. However, when it infects a cell with the trans-activating noncoding RNA, the cell should lyse because the RBS of the developmental is no longer sequestered by the cis-repressed RNA loop. | |
==Riboregulator Design== | ==Riboregulator Design== | ||
| - | + | ===Design Background=== | |
| - | + | ||
Our riboregulators consist of two interacting parts: cis and trans. The cis sequence is located downstream from the promoter to a gene of interest and upstream of the ribosome binding site (RBS). This cis sequence is complementary to the RBS and forms a stem-loop at the 5’ end of the mRNA after transcription, thus blocking ribosome binding and translation. Gene expression is turned on with a trans noncoding RNA. This trans sequence is produced from another promoter in a different plasmid and targets the cis-repressed RNA with high specificity. When the trans transcript is available, it binds to the cis region and thus opens the RBS for ribosome docking and subsequent translation. | Our riboregulators consist of two interacting parts: cis and trans. The cis sequence is located downstream from the promoter to a gene of interest and upstream of the ribosome binding site (RBS). This cis sequence is complementary to the RBS and forms a stem-loop at the 5’ end of the mRNA after transcription, thus blocking ribosome binding and translation. Gene expression is turned on with a trans noncoding RNA. This trans sequence is produced from another promoter in a different plasmid and targets the cis-repressed RNA with high specificity. When the trans transcript is available, it binds to the cis region and thus opens the RBS for ribosome docking and subsequent translation. | ||
| + | |||
The cis repressive sequences are approximately 50 base pairs and do not alter the reading frame of the native gene. The trans RNA is approximately 90 base pairs and contains a nucleotide sequence that is complementary to the cis RNA. RNAstructure3 software was used to predict the secondary structure as well as the free energy of the cis and trans elements and their interactions. | The cis repressive sequences are approximately 50 base pairs and do not alter the reading frame of the native gene. The trans RNA is approximately 90 base pairs and contains a nucleotide sequence that is complementary to the cis RNA. RNAstructure3 software was used to predict the secondary structure as well as the free energy of the cis and trans elements and their interactions. | ||
| + | [[Image:Cis trans diagram.JPG|center|frame|500px|Figure adopted from: Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, and Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol 2004 Jul; 22(7) 841-7.]] | ||
| - | |||
| + | ===Cis Repression=== | ||
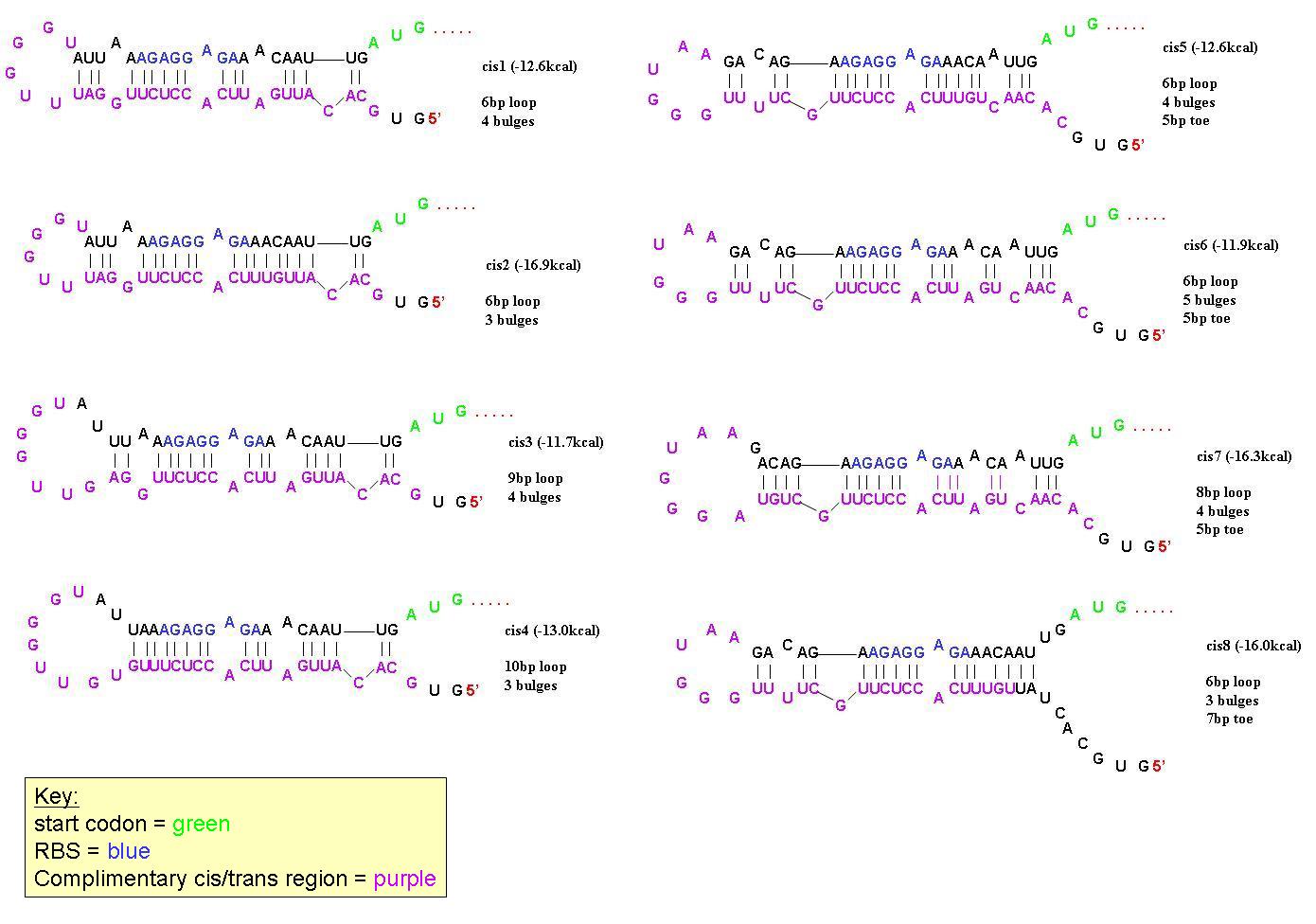
| + | In order to find a riboregulator with the necessary dynamic range, a total of eight cis repressive elements were designed (cr1-cr8). These consisted of the same overall stem-loop structure, but had varying strengths of complementarity within the stem, as seen by their free energies. The designs of cr1-cr4 were based on previous work done by Collins et. al. and those for cr5-cr8 were based on work done by the University of California Berkeley 2006 International Genetically Engineered Machines team. | ||
| - | + | In order to determine the optimal level of repression, various aspects of RNA secondary structure were considered, including inner loops, single base pair bulges, and varying loop sizes. Higher free energies (i.e. less complementarity due to base pair mismatches) favor activation by taRNA because they destabilize the stem and facilitate the open RBS form upon addition of taRNA. The cis elements were inserted downstream of the Ptet promoter. The stem consisted of approximately 20-nt and the loop ranged from 6 to 10-nt. The YFP gene was inserted directly downstream of the cis sequence. Flow cytometry measurements were taken to quantify the expression of the cis riboregulated YFP gene. | |
| - | + | [[Image:Cis designs 1.JPG|center|thumb|700px|All 8 cis-repressive riboregulators have similar structure but differ in the number of base-pairs in end-loops, number of bulges, and number of base pairs on the 5' toe. They were designed this way to find the optimal dynamic range in repression/activation of protein expression.]] | |
| - | + | ||
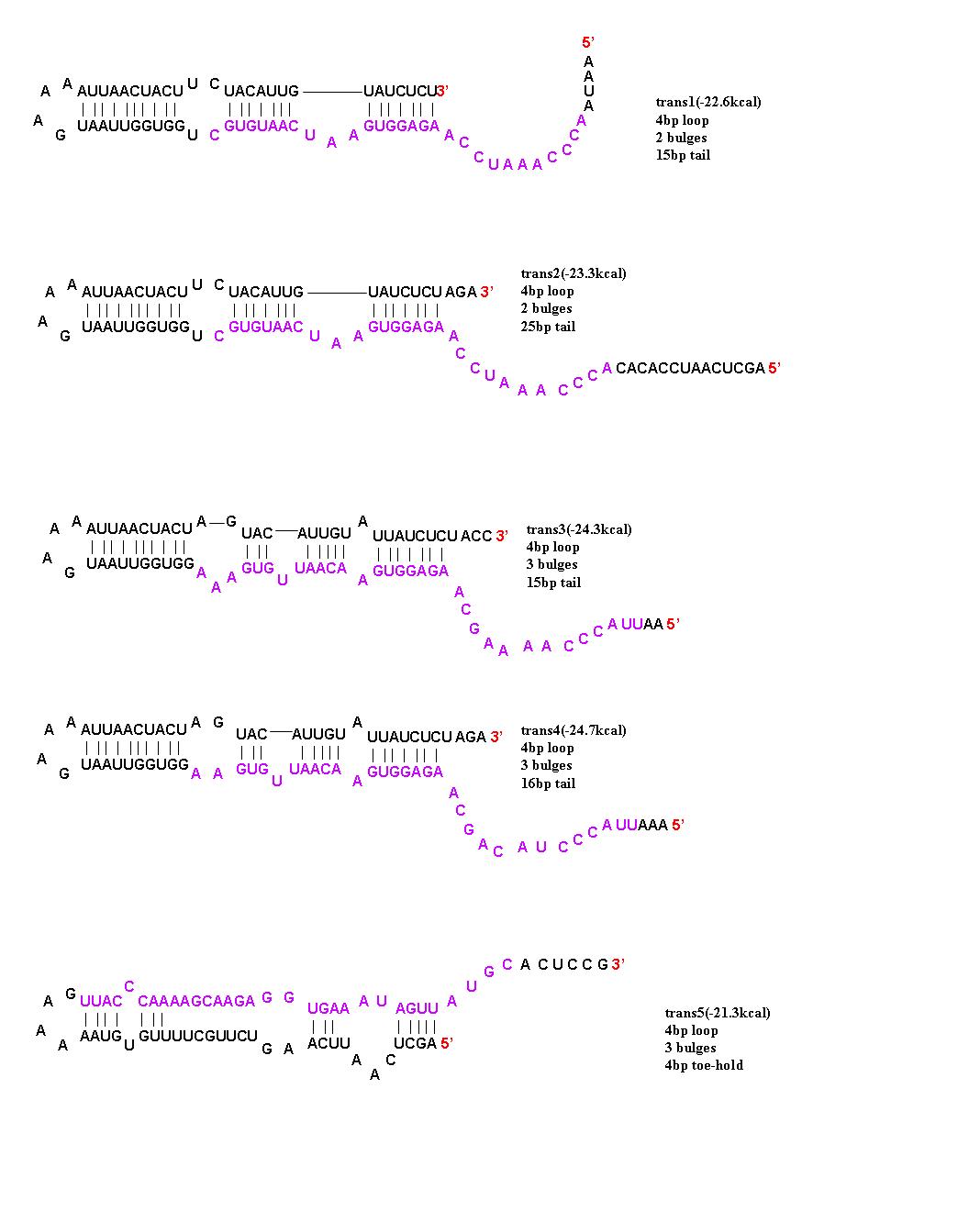
| + | ===Trans Activation=== | ||
| + | To initiate translation, five trans-activating elements (ta1-ta5) were designed. Each sequence binds to the cis-repressive elements and opens up the RBS. For ta1-ta4, part of the sequence was complementary to the hairpin loop of the cr element and extending in the 5’ direction. Element ta5 was designed to bind to the 5’ end of the cis regulator and open up the hairpin in the 3’ direction. trans1 and trans2 are complimentary to cis1-cis4; trans3 is complimentary to cis5, cis6, and cis8; trans4 is complimentary to cis7; and trans5 is complimentary to cis5-8. The activation with these ta elements is currently being [[Caltech/Project/Riboregulator_Results|determined]]. | ||
| - | + | [[Image:Trans designs 1.JPG|center|thumb|700px|The 5 trans-activating riboregulators are shown with purple bases indicating regions of complimentarity to the cis-repressive riboregulators.]] | |
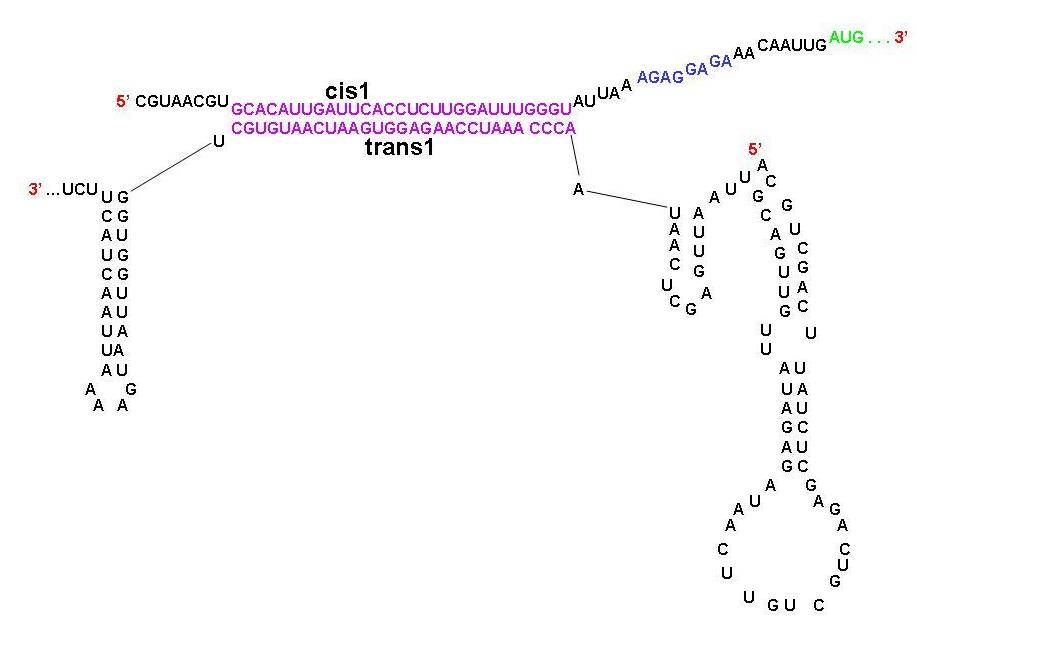
| - | + | ===Cis-trans combinations=== | |
| - | + | [[Image:Cis trans compliments.JPG|center|thumb|700px|A schematic diagram showing the parts of cis1 repressor and trans1 activator that are complimentary to each other. Note that the RBS is now open.]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [[Image:Cis trans compliments.JPG| | + | |
==Experiments with Riboregulators== | ==Experiments with Riboregulators== | ||
| - | In order to have a working system in which phage infection outcome is controlled, the expression of genes under the control of a riboregulator must be determined. | + | In order to have a working system in which phage infection outcome is controlled, the expression of genes under the control of a riboregulator must be determined. Then, once we determine the amount of N, Q, or Cro necessary for infection, an appropriate riboregulator can be integrated into the system. |
Before combining the phage protein expression with the riboregulator part of the system, it is necessary to engineer various riboregulators and determine the percent of protein (yellow fluorescent protein or YFP) in the presence and absence of trans RNA. By quantifying the riboregulated YFP system, we could then apply the results to each protein. | Before combining the phage protein expression with the riboregulator part of the system, it is necessary to engineer various riboregulators and determine the percent of protein (yellow fluorescent protein or YFP) in the presence and absence of trans RNA. By quantifying the riboregulated YFP system, we could then apply the results to each protein. | ||
==Status and Future Plans== | ==Status and Future Plans== | ||
| + | Please see our [[Caltech/Project/Riboregulator_Results|results page]] for the current status of these experiments. | ||
| - | |||
| - | |||
| - | |||
|} | |} | ||
</div> | </div> | ||
Latest revision as of 03:46, 27 October 2007
iGEM 2007
Home
Highlights
Project
People
Protocols
|