Chiba/Flagella/Making FliC-his
From 2007.igem.org
< Chiba(Difference between revisions)
| (2 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
*ExSite PCR | *ExSite PCR | ||
*design primer | *design primer | ||
| - | *Among the code of FliC, we selected five fragment codes from the outside part of the target filament, then designed each primers(207,228,240,252,267). Mounted pUC19-FliC to pUC19-FliC-vector. | + | *Among the code of FliC, we selected five fragment codes from the outside part of the target filament, then designed each primers(207,228,240,252,267). Mounted pUC19-FliC to pUC19-FliC-vector.<br><br> |
*[Reaction Conditions] | *[Reaction Conditions] | ||
**Template 1μL | **Template 1μL | ||
| Line 35: | Line 35: | ||
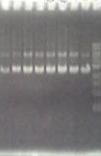
| - | + | [[Image:Gel_check.JPG]]<br> | |
| - | [[Image:Gel_check.JPG]] | + | Fig. GelCheck showing pUC19-FliC-vector(3973bp). |
Latest revision as of 04:52, 27 October 2007
- fliC is inserted in pUC19 vector→pUC19-FliC
- ExSite PCR
- design insert(6×Histidine-Tag)
- Lagation pUC19-FliC vector and insert(flic+6×His-tag)
- GelCheck
- ExSite PCR
- design primer
- Among the code of FliC, we selected five fragment codes from the outside part of the target filament, then designed each primers(207,228,240,252,267). Mounted pUC19-FliC to pUC19-FliC-vector.
- [Reaction Conditions]
- Template 1μL
- Primer(Foward and Reverse) 5μL
- 10×buffer 10μL
- dNTPmix 10μL
- vent 1μL
- dH2O 68μL
- design insert
- six histidine loop peptide ("His-loop")
- -Ser-Ser-His-His-His-His-His-His-Ser-Ser
- encoded by the oligo 5'-TCT-TCT-CAT-CAT-CAT-CAT-CAT-CAT-TCC-TCC-3'
- and anti oligo 5'-GGA-GGA-ATG-ATG-ATG-ATG-ATG-ATG-AGA-AGA-3'
- restriction site (N termimal: EcoRⅠ,C terminal: NcoⅠ
- Ligation pUC19-FliC vector with insert.
- [Reaction Conditions]
- insert 2μL
- pUC19-FliC vector 2μL
- ×5 Ligase Buffer 4μL
- Ligase 1μL
- dH2O 11μL
Fig. GelCheck showing pUC19-FliC-vector(3973bp).