ETHZ
From 2007.igem.org
(→'''.:: Site Map ::.''') |
Stefan Luzi (Talk | contribs) (→Multipurpose Cell Lines) |
||
| (113 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:ETHZ_banner.png|830px]] | |
| + | <!-- | ||
| + | <center>[[ETHZ | Main Page]] [[ETHZ/Model | System Modeling]] [[ETHZ/Simulation | Simulations]] [[ETHZ/Biology | System Implementation]] [[ETHZ/Biology/Lab| Lab Notes]] [[ETHZ/Meet_the_team | Meet the Team]] [[ETHZ/Internal | Team Notes]] [[ETHZ/Pictures | Pictures!]]</center><br> | ||
| + | --> | ||
| + | __NOTOC__ | ||
| + | <html> | ||
| + | <script type="text/javascript" src="http://christos.bergeles.net/eth_dropdowntabs.js"> | ||
| + | |||
| + | /*********************************************** | ||
| + | * Drop Down Tabs Menu- (c) Dynamic Drive DHTML code library (www.dynamicdrive.com) | ||
| + | * This notice MUST stay intact for legal use | ||
| + | * Visit Dynamic Drive at http://www.dynamicdrive.com/ for full source code | ||
| + | ***********************************************/ | ||
| + | |||
| + | </script> | ||
| + | |||
| + | <!-- CSS for Drop Down Tabs Menu #1 --> | ||
| + | <link rel="stylesheet" type="text/css" href="http://christos.bergeles.net/eth_ddcolortabs.css" /> | ||
| + | <div id="colortab" class="ddcolortabs"> | ||
| + | <ul> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ" title="Home" rel="dropmenu_home"><span>Home</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model" title="Modeling" rel="dropmenu_modeling"><span>System Modeling</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation" title="Simulations" rel="dropmenu_simulation"><span>Simulations</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology" title="System Implementation" rel="dropmenu_biology"><span>System Implementation</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team" title="Meet the team" rel="dropmenu_meettheteam"><span>Meet the team</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Pictures" title="Pictures!" rel="dropmenu_pictures"><span>Pictures!</span></a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div class="ddcolortabsline"> </div> | ||
| + | |||
| + | |||
| + | <!--1st drop down menu --> | ||
| + | <div id="dropmenu_home" class="dropmenudiv_a"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Team_Members">Team Members</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Acknowledgments">Acknowledgments</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Site_Map">Site map</a> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <!--2nd drop down menu --> | ||
| + | <div id="dropmenu_modeling" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Model_Overview">Model Overview</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Detailed_Model">Detailed Model</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Final_Model">Final Model</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Modeling_Basics">Modeling Basics Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Mathematical_Model">Mathematical Model</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FSM">FSM View Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FlipFlop">Flip-Flop View Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Parameters">Parameters Page</a> | ||
| + | </div> | ||
| + | |||
| + | <!--3rd drop down menu --> | ||
| + | <div id="dropmenu_simulation" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Simulation_of_Test_Cases">Test Cases</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Sensitivity_Analysis">Sensitivity Analysis</a> | ||
| + | </div> | ||
| + | |||
| + | <!--4th drop down menu --> | ||
| + | <div id="dropmenu_biology" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#The_Complete_System">The Complete System</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#System_Phases">System Phases</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#Current_Cloning_Status">Current Cloning Status</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/parts">System Parts Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/Lab">Lab Notes Page</a> | ||
| + | </div> | ||
| + | |||
| + | <!--5th drop down menu --> | ||
| + | <div id="dropmenu_meettheteam" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#The_ETH_Zurich_07_Team">The ETH Zurich 07 Team</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#Team_Description">Team Description</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Internal">Brainstorming Page</a> | ||
| + | </div> | ||
| - | < | + | <script type="text/javascript"> |
| + | //SYNTAX: tabdropdown.init("menu_id", [integer OR "auto"]) | ||
| + | tabdropdown.init("colortab", 3) | ||
| + | </script> | ||
| + | </html> | ||
__NOTOC__ | __NOTOC__ | ||
| - | <center><font size = | + | <html> |
| + | <p align="center"><font size="6"><b>ETH Zurich - educatETH <i>E.coli</i> System</b></font></p> | ||
| + | </body> | ||
| + | </html> | ||
=Introduction= | =Introduction= | ||
| - | <blockquote>" | + | <blockquote>"All <i>E.coli</i> 's are equal, but some <i>E.coli</i> 's are more equal than others..." ''(freely adapted from "Animal Farm" by George Orwell)''</blockquote> |
| - | + | ... this is what George Orwell would have written, were he a synthetic biologist. In the <i>E.coli</i> colonies on petri dishes, all bacteria are equal; except for some special ones. Our project is about designing such special <i>E.coli</i> that are "more equal" than the rest: they have the ability to be trained in order to memorize and recognize their environment in the future. Their story will be presented through this wiki ... | |
| - | + | ==Motivation== | |
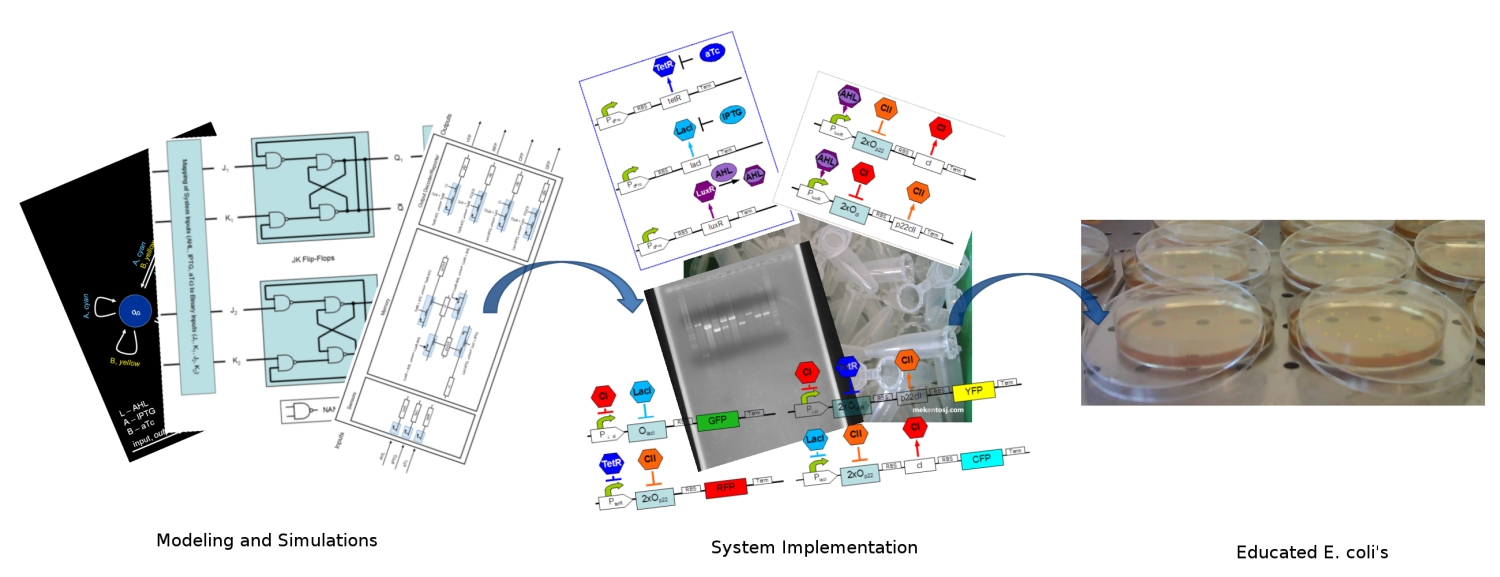
| - | + | [[Image:ethz_main_pic.jpg|right|thumb|<b>Fig. 1</b>: Artist's approach to the different stages of the development. We started by modeling and simulating the system. We continued by specifying the DNA strands for its implementation. Finally in the end, our system should report with different fluorescent proteins (image edited)|600px]] | |
| - | + | Our combined team of biologists and engineers is coping with the problem of implementing memory capabilities in bacterial colonies. First, ''E.coli'' are able to respond differently (with distinct fluorescent proteins) to two different inputs (we used chemicals). Second, they remember which input was presented to them. Finally, when confronted with a new input, they are able to recognize whether it is the one that they were trained with or not. | |
| - | + | In other words, in this project '''we are educating the <i>E.coli</i>'''! | |
| - | The ETH Zurich team consists of good mixture between biologists and engineering students | + | ===Multipurpose Cell Lines=== |
| + | |||
| + | Our system has the ability to behave in different ways according to an internal toggle inside it switching states based on the chemical substances that the system is exposed to. The toggle states could generally be used to trigger events such as enzyme synthesis, transcriptional regulation, virion production, or even cell death. Therefore, one may view the bacterial cell line containing this system as a multipurpose cell line. By adding a certain chemical to a cell line, the latter may be trained to exhibit a desired behavior, and then it is not necessary any more to construct two independent cell lines. | ||
| + | |||
| + | This means that one applies an “input engineering” instead of a “DNA engineering” approach. If one extends this idea to several multi-inducible toggle switches being harbored in the same cell line, the number of possible phenotypes increases to 2<i><sup>n</sup></i>, where <i>n</i> equals the number of toggle switches. For example, if one would have 5 such toggle switches inside a cell line, 32 different behavior patterns would be possible. | ||
| + | |||
| + | For the purpose of creating a toggle switch that is activated in a specific phase only and not always (a multi-inducible toggle switch), as is required for stable biological automatons, we introduced the concept of [https://2007.igem.org/ETHZ/Biology/parts#double_promoters double promoters] to the [http://partsregistry.org/Main_Page Registry of Standard Biological Parts], which can be helpful for engineering systems which exhibit a desired behavior only at specific times. | ||
| + | |||
| + | ===Link to Epigenetics=== | ||
| + | Epigenetics refers to features like chromatin or DNA modifications that do not involve changes in the underlying DNA sequence and are stable over many cell divisions [1],[2]. If one has a closer look at our proposed system, one can also view it as a model-system for epigenetics: Although the DNA sequence itself stays the same, two different subpopulations of cells with different phenotypes can develop from it. Put simply, depending in which state (subpopulation) the toggle switch is, the cells will produce different fluorescent proteins upon addition of two different inducer molecules. Therefore, the epigenetic feature here is the binding of specific repressor proteins whose production is dependent on the toggle switch state. | ||
| + | |||
| + | ===Intelligent Biosensors and Self-Adaptation=== | ||
| + | |||
| + | The system is capable of sensing different chemicals and producing different fluorescent proteins. Since the cells can be trained to produce one of several specific fluorescent protein types when a certain chemical is present, one can also view those cells as intelligent biosensors which recognize chemical substances according to a training phase. The intelligent biosensors are not limited to detect chemicals; temperature, pH, light, pressure could be detected with an appropriate system as well. Such an application could be especially of interest when the environment to be probed is harmful for humans, for example due to high toxicity. | ||
| + | |||
| + | ==Team Members== | ||
| + | [[Image:ETHZ_Group_photo_6.png|right|thumb|The ETH Zurich iGEM2007 Team|270px]] | ||
| + | |||
| + | The ETH Zurich team consists of a good mixture between biologists and engineering students. | ||
| + | We are: | ||
<ul> | <ul> | ||
| - | <li><i>Undergraduate students</i>: | + | <li><i>Undergraduate students</i>: <br> |
| - | [https://2007.igem.org/User:brutsche Martin Brutsche], [https://2007.igem.org/User:kdikaiou Katerina Dikaiou], [https://2007.igem.org/User:Raphael Raphael Guebeli], [https://2007.igem.org/User:hoehnels Sylke Hoehnel], | + | [https://2007.igem.org/User:brutsche Martin Brutsche], [https://2007.igem.org/User:kdikaiou Katerina Dikaiou], <br>[https://2007.igem.org/User:Raphael Raphael Guebeli], [https://2007.igem.org/User:hoehnels Sylke Hoehnel], <br> |
[https://2007.igem.org/Nan_Li Nan Li], | [https://2007.igem.org/Nan_Li Nan Li], | ||
[https://2007.igem.org/User:Stefan Stefan Luzi] | [https://2007.igem.org/User:Stefan Stefan Luzi] | ||
| - | <li><i>Graduate students</i>: | + | <li><i>Graduate students</i>: <br> |
[http://christos.bergeles.net Christos Bergeles], | [http://christos.bergeles.net Christos Bergeles], | ||
| - | [http://www.tik.ee.ethz.ch/~sop/people/thohm/ Tim Hohm], [http://www.fussenegger.ethz.ch/people/kemmerc Christian Kemmer], | + | [http://www.tik.ee.ethz.ch/~sop/people/thohm/ Tim Hohm], <br> |
| - | [https://2007.igem.org/User:JoeKnight Joseph Knight], | + | [http://www.fussenegger.ethz.ch/people/kemmerc Christian Kemmer], |
| - | [http://csb.inf.ethz.ch/people/uhr.html Markus Uhr | + | [https://2007.igem.org/User:JoeKnight Joseph Knight], <br> |
| - | + | [http://www.ricomoeckel.de Rico Möckel], | |
| - | <li><i>Project advisors</i>: | + | [http://csb.inf.ethz.ch/people/uhr.html Markus Uhr] |
| - | [http://www.ipe.ethz.ch/laboratories/bpl/people/panke Sven Panke], | + | <li><i>Project advisors</i>: <br> |
| + | [http://www.ipe.ethz.ch/laboratories/bpl/people/panke Sven Panke], <br> | ||
[http://csb.inf.ethz.ch/people/stelling.html Joerg Stelling] | [http://csb.inf.ethz.ch/people/stelling.html Joerg Stelling] | ||
</ul> | </ul> | ||
| + | |||
For more information about us, visit our [[ETHZ/Meet_the_team | Meet the Team]] page. | For more information about us, visit our [[ETHZ/Meet_the_team | Meet the Team]] page. | ||
| - | = | + | ==Acknowledgments== |
| - | The idea for the project as well as its implementation was done by the ETH iGEM 2007 team. | + | The idea for the project as well as its implementation was done solely by the ETH iGEM 2007 team. We would like to thank the people in [http://www.ipe.ethz.ch/laboratories/bpl/index Sven Panke's Lab], especially Andreas Meyer who was always there for us when we had a problem. Additionally, we would like to thank [http://www.facs.ethz.ch Alfredo Franco-Obregón's lab] and Oralea Büchi for the help with the flow cytometry. |
| - | We would also like to acknowledge the | + | We would also like to acknowledge the financial support by [http://europa.eu EU], the [http://www.ethz.ch ETH Zurich], and [http://www.geneart.com GeneArt]: |
<center> | <center> | ||
{| border="0" | {| border="0" | ||
| Line 53: | Line 156: | ||
</center> | </center> | ||
| - | =Site Map= | + | ==Site Map== |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | In this wiki, we will present to you a detailed description of the proposed system: starting with the [[ETHZ/Model | modeling of the system]], we describe both, [[ETHZ/Simulation | simulations and theoretical considerations]] of the system, as well as the actual [[ETHZ/Biology | implementation using bio-bricks]] accompanied by our [[ETHZ/Biology/Lab | lab notes]]. Additionally, you will find further [[ETHZ/Meet_the_team | information on the team]], more details about [[ETHZ/Internal | ideas we developed]] before we came up with the system we finally implemented, and some [[ETHZ/Pictures | pictures]] documenting our work. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | The site map of our wiki is the following: | |
| - | + | ||
| - | + | {| class="wikitable" width="100%" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | |
| - | + | !width="34%"| Modeling Pages | |
| - | + | !width="22%"| Biology Pages | |
| - | + | !width="22%"| ETHZ Team Pages | |
| - | + | !width="22%"| Links | |
| - | + | |- | |
| - | + | | [[ETHZ/Model | Modeling of the learning system]] | |
| - | + | | [[ETHZ/Biology | Biological implementation]] | |
| - | + | | [[ETHZ/Meet_the_team | Team page]] | |
| - | + | | [https://2006.igem.org/wiki/index.php/ETH_Zurich_2005 The ETH Zurich 2005 project] | |
| - | + | |- | |
| - | + | | [[ETHZ/FlipFlop | Representation using flip-flops]] | |
| - | + | | [[ETHZ/Biology/parts | Biobricks/parts]] | |
| - | + | | [[ETHZ/Pictures | Pictures]] | |
| - | + | | [https://2006.igem.org/wiki/index.php/ETH_Zurich_2006 The ETH Zurich 2006 project] | |
| - | + | |- | |
| - | + | | [[ETHZ/FSM | Representation using finite state machines]] | |
| - | + | | [[ETHZ/Biology/Lab | Lab notes]] | |
| - | + | | [[ETHZ/Internal | Brainstorming sessions]] | |
| - | + | | | |
| - | + | |- | |
| - | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | + | | [[ETHZ/Simulation | Model simulations and theoretical considerations]] |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | |- | |
| - | + | | [[ETHZ/Parameters | Parameters used in our simulations]] | |
| - | + | | | |
| - | + | | | |
| - | + | | | |
| - | + | |- | |
| - | + | |} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | [ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | == References == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [http://www.nature.com/nature/journal/v447/n7143/abs/nature05913.html;jsessionid=62903C604764B175945C03DB8639ECBD [1] Bird A] <i>"Perceptions of epigenetics"</i>, Nature 447:396-398, 2007 <br /> | |
| - | + | [http://linkinghub.elsevier.com/retrieve/pii/S096098220701007X [2] Ptashne M] <i>"On the use of the word ‘epigenetic’"</i>, Current Biology 17(7):R233-R236, 2007 <br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | <html><body> | + | <html><body> |
| - | <a> | + | <a href="http://www3.clustrmaps.com/counter/maps.php?url=https://2007.igem.org/ETHZ" id="clustrMapsLink"><img src="http://www3.clustrmaps.com/counter/index2.php?url=https://2007.igem.org/ETHZ" style="border:0px;" alt="Locations of visitors to this page" title="Locations of visitors to this page" id="clustrMapsImg" onError="this.onError=null; this.src='http://www2.clustrmaps.com/images/clustrmaps-back-soon.jpg'; document.getElementById('clustrMapsLink').href='http://www2.clustrmaps.com'" /> |
| - | <img src="http://www.easycounter.com/counter.php?stefanluzi" | + | </a> |
| - | border="0" alt="Welcome to ETHz - iGEM07"></a> | + | |
| + | <html><body> | ||
| + | <a> | ||
| + | <img src="http://www.easycounter.com/counter.php?stefanluzi" | ||
| + | border="0" alt="Welcome to ETHz - iGEM07"></a> | ||
</body></html> | </body></html> | ||
Latest revision as of 20:20, 26 October 2007
ETH Zurich - educatETH E.coli System