ETHZ/FlipFlop
From 2007.igem.org
(→'''.:: An Engineer's View on Biological Learning ::.''') |
|||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:ETHZ_banner.png|830px]] | |
| - | [[Image: | + | <!-- |
| - | + | <center>[[ETHZ | Main Page]] [[ETHZ/Model | System Modeling]] [[ETHZ/Simulation | Simulations]] [[ETHZ/Biology | System Implementation]] [[ETHZ/Biology/Lab| Lab Notes]] [[ETHZ/Meet_the_team | Meet the Team]] [[ETHZ/Internal | Team Notes]] [[ETHZ/Pictures | Pictures!]]</center><br> | |
| + | --> | ||
| + | __NOTOC__ | ||
| + | <html> | ||
| + | <script type="text/javascript" src="http://christos.bergeles.net/eth_dropdowntabs.js"> | ||
| - | + | /*********************************************** | |
| + | * Drop Down Tabs Menu- (c) Dynamic Drive DHTML code library (www.dynamicdrive.com) | ||
| + | * This notice MUST stay intact for legal use | ||
| + | * Visit Dynamic Drive at http://www.dynamicdrive.com/ for full source code | ||
| + | ***********************************************/ | ||
| + | </script> | ||
| + | |||
| + | <!-- CSS for Drop Down Tabs Menu #1 --> | ||
| + | <link rel="stylesheet" type="text/css" href="http://christos.bergeles.net/eth_ddcolortabs.css" /> | ||
| + | <div id="colortab" class="ddcolortabs"> | ||
| + | <ul> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ" title="Home" rel="dropmenu_home"><span>Home</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model" title="Modeling" rel="dropmenu_modeling"><span>System Modeling</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation" title="Simulations" rel="dropmenu_simulation"><span>Simulations</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology" title="System Implementation" rel="dropmenu_biology"><span>System Implementation</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team" title="Meet the team" rel="dropmenu_meettheteam"><span>Meet the team</span></a></li> | ||
| + | <li><a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Pictures" title="Pictures!" rel="dropmenu_pictures"><span>Pictures!</span></a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | <div class="ddcolortabsline"> </div> | ||
| + | |||
| + | |||
| + | <!--1st drop down menu --> | ||
| + | <div id="dropmenu_home" class="dropmenudiv_a"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Team_Members">Team Members</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Acknowledgments">Acknowledgments</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ#Site_Map">Site map</a> | ||
| + | </div> | ||
| + | |||
| + | |||
| + | <!--2nd drop down menu --> | ||
| + | <div id="dropmenu_modeling" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Model_Overview">Model Overview</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Detailed_Model">Detailed Model</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Final_Model">Final Model</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Modeling_Basics">Modeling Basics Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Model#Mathematical_Model">Mathematical Model</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FSM">FSM View Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/FlipFlop">Flip-Flop View Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Parameters">Parameters Page</a> | ||
| + | </div> | ||
| + | |||
| + | <!--3rd drop down menu --> | ||
| + | <div id="dropmenu_simulation" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Simulation_of_Test_Cases">Test Cases</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Simulation#Sensitivity_Analysis">Sensitivity Analysis</a> | ||
| + | </div> | ||
| + | |||
| + | <!--4th drop down menu --> | ||
| + | <div id="dropmenu_biology" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#Introduction">Introduction</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#The_Complete_System">The Complete System</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#System_Phases">System Phases</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology#Current_Cloning_Status">Current Cloning Status</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/parts">System Parts Page</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Biology/Lab">Lab Notes Page</a> | ||
| + | </div> | ||
| + | |||
| + | <!--5th drop down menu --> | ||
| + | <div id="dropmenu_meettheteam" class="dropmenudiv_a" style="width: 150px;"> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#The_ETH_Zurich_07_Team">The ETH Zurich 07 Team</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Meet_the_team#Team_Description">Team Description</a> | ||
| + | <a href="https://2007.igem.org/wiki/index.php?title=ETHZ/Internal">Brainstorming Page</a> | ||
| + | </div> | ||
| + | |||
| + | <script type="text/javascript"> | ||
| + | //SYNTAX: tabdropdown.init("menu_id", [integer OR "auto"]) | ||
| + | tabdropdown.init("colortab", 3) | ||
| + | </script> | ||
| + | |||
| + | </html> | ||
| + | |||
| + | __NOTOC__ | ||
| + | |||
| + | =An Engineer's View on Biological Learning= | ||
| + | |||
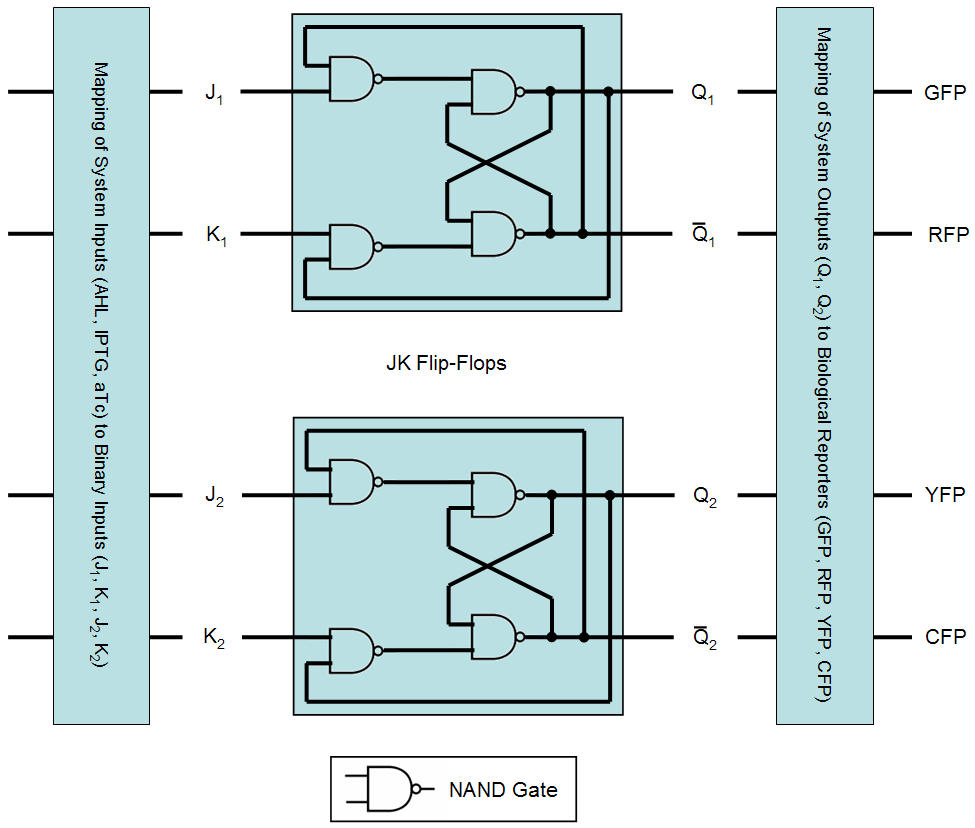
| + | [[Image:FlipFlop.png|thumb|450px|<b>Fig. 1</b>: Schematic view on the system implemented by logical gates.]] | ||
| + | An equivalent system behavior can be realized using [http://en.wikipedia.org/wiki/Flip-flop_%28electronics%29 flip-flops], implemented by [http://en.wikipedia.org/wiki/Logic_gate logical gates]: | ||
| + | |||
| + | As already shown in the [[ETHZ/FSM | finite state machine representation]] (FSM), the proposed system consists of three different states and recognizes four different inputs. In total, this adds up to twelve different transitions. In order to implement these twelve transitions with a digital system, at least four binary inputs are required (allowing for a maximum of 16 transitions) and since the proposed system has four different outputs, at least two binary outputs are required. Here, we chose an implementation using two JK flip-flops consisting of [http://en.wikipedia.org/wiki/Nand_gate#NAND_gate NAND gates]. An overview of the resulting system is given in Fig. 1 and details concerning flip-flop and NAND gate behavior are given in the following table: | ||
{| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| - | | A | + | ! J !! K !! ''Q''<sub>next</sub> !! width="50" | !! A !! B !! A NAND B |
|- align="center" | |- align="center" | ||
| - | |0 || 0 || 0 || 1 | + | | 0 || 0 || ''Q''<sub>prev</sub> || || 0 || 0 || 1 |
|- align="center" | |- align="center" | ||
| - | |0 || 1 || 0 || 1 | + | | 0 || 1 || 0 || || 0 || 1 || 1 |
|- align="center" | |- align="center" | ||
| - | |1 || 0 || 0 || 1 | + | | 1 || 0 || 1 || || 1 || 0 || 1 |
| + | |- align="center" | ||
| + | | 1 || 1 || [http://en.wikipedia.org/wiki/Inverse_%28logic%29 ¬''Q''<sub>prev</sub>] || || 1 || 1 || 0 | ||
| + | |} | ||
| + | |||
| + | When initializing the system we set both outputs ''Q'' to zero. The mapping from inputs ([[ETHZ/FSM | AHL+IPTG, IPTG, AHL+aTc, aTc]]) and system states ([[ETHZ/FSM | ''q''<sub>0</sub>, ''q''<sub>1</sub>, ''q''<sub>2</sub>]]) to the binary inputs (J<sub>1</sub>, K<sub>1</sub>, J<sub>2</sub>, K<sub>2</sub>) is given by following table: | ||
| + | |||
| + | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| + | !states/inputs !! AHL+IPTG !! IPTG !! AHL+aTc !! aTc | ||
| + | |- align="center" | ||
| + | ! ''q''<sub>0</sub> | ||
| + | |J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=1, K<sub>2</sub>=0 | ||
| + | |J<sub>1</sub>=0, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | |J<sub>1</sub>=1, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | |J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | |- align="center" | ||
| + | ! ''q''<sub>1</sub> | ||
| + | |J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=1, K<sub>2</sub>=0 | ||
| + | |J<sub>1</sub>=0, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | |J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=0, K<sub>2</sub>=0 | ||
| + | |J<sub>1</sub>=0, K<sub>1</sub>=0, J<sub>2</sub>=1, K<sub>2</sub>=1 | ||
| + | |- align="center" | ||
| + | ! ''q''<sub>2</sub> | ||
| + | |J<sub>1</sub>=1, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=0 | ||
| + | |J<sub>1</sub>=1, K<sub>1</sub>=1, J<sub>2</sub>=0, K<sub>2</sub>=0 | ||
| + | |J<sub>1</sub>=1, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | |J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | |} | ||
| + | |||
| + | Here, we assume that the [[ETHZ/FSM | FSM system states]] ''q''<sub>0</sub>, ''q''<sub>1</sub>, ''q''<sub>2</sub> are mapped to the outputs ''Q''<sub>1</sub> and ''Q''<sub>2</sub> as follows: | ||
| + | |||
| + | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| + | !FSM states/output coding !! ''Q''<sub>1</sub> !! ''Q''<sub>2</sub> | ||
| + | |- align="center" | ||
| + | !''q''<sub>0</sub>||0||0 | ||
| + | |- align="center" | ||
| + | !''q''<sub>0</sub>||0||1 | ||
| + | |- align="center" | ||
| + | !''q''<sub>0</sub>||1||0 | ||
| + | |} | ||
| + | |||
| + | The final mapping from transitions to the biological reporters is then given by: | ||
| + | {| class="wikitable" border="1" cellspacing="0" cellpadding="2" style="text-align:left; margin: 1em 1em 1em 0; background: #f9f9f9; border: 1px #aaa solid; border-collapse: collapse;" | ||
| + | !J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=1, K<sub>2</sub>=0 | ||
| + | !J<sub>1</sub>=0, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | !J<sub>1</sub>=1, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | !J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | !J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=1, K<sub>2</sub>=0 | ||
| + | !J<sub>1</sub>=0, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | !J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=0, K<sub>2</sub>=0 | ||
| + | !J<sub>1</sub>=0, K<sub>1</sub>=0, J<sub>2</sub>=1, K<sub>2</sub>=1 | ||
| + | !J<sub>1</sub>=1, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=0 | ||
| + | !J<sub>1</sub>=1, K<sub>1</sub>=1, J<sub>2</sub>=0, K<sub>2</sub>=0 | ||
| + | !J<sub>1</sub>=1, K<sub>1</sub>=0, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
| + | !J<sub>1</sub>=0, K<sub>1</sub>=1, J<sub>2</sub>=0, K<sub>2</sub>=1 | ||
|- align="center" | |- align="center" | ||
| - | | | + | | ''cyan'' |
| + | | ''cyan'' | ||
| + | | ''red'' | ||
| + | | ''cyan'' | ||
| + | | ''cyan'' | ||
| + | | ''cyan'' | ||
| + | | ''yellow'' | ||
| + | | ''green'' | ||
| + | | ''cyan'' | ||
| + | | ''cyan'' | ||
| + | | ''cyan'' | ||
| + | | ''yellow'' | ||
|} | |} | ||
Latest revision as of 14:27, 26 October 2007
An Engineer's View on Biological Learning
An equivalent system behavior can be realized using flip-flops, implemented by logical gates:
As already shown in the finite state machine representation (FSM), the proposed system consists of three different states and recognizes four different inputs. In total, this adds up to twelve different transitions. In order to implement these twelve transitions with a digital system, at least four binary inputs are required (allowing for a maximum of 16 transitions) and since the proposed system has four different outputs, at least two binary outputs are required. Here, we chose an implementation using two JK flip-flops consisting of NAND gates. An overview of the resulting system is given in Fig. 1 and details concerning flip-flop and NAND gate behavior are given in the following table:
| J | K | Qnext | A | B | A NAND B | |
|---|---|---|---|---|---|---|
| 0 | 0 | Qprev | 0 | 0 | 1 | |
| 0 | 1 | 0 | 0 | 1 | 1 | |
| 1 | 0 | 1 | 1 | 0 | 1 | |
| 1 | 1 | ¬Qprev | 1 | 1 | 0 |
When initializing the system we set both outputs Q to zero. The mapping from inputs ( AHL+IPTG, IPTG, AHL+aTc, aTc) and system states ( q0, q1, q2) to the binary inputs (J1, K1, J2, K2) is given by following table:
| states/inputs | AHL+IPTG | IPTG | AHL+aTc | aTc |
|---|---|---|---|---|
| q0 | J1=0, K1=1, J2=1, K2=0 | J1=0, K1=0, J2=0, K2=1 | J1=1, K1=0, J2=0, K2=1 | J1=0, K1=1, J2=0, K2=1 |
| q1 | J1=0, K1=1, J2=1, K2=0 | J1=0, K1=0, J2=0, K2=1 | J1=0, K1=1, J2=0, K2=0 | J1=0, K1=0, J2=1, K2=1 |
| q2 | J1=1, K1=0, J2=0, K2=0 | J1=1, K1=1, J2=0, K2=0 | J1=1, K1=0, J2=0, K2=1 | J1=0, K1=1, J2=0, K2=1 |
Here, we assume that the FSM system states q0, q1, q2 are mapped to the outputs Q1 and Q2 as follows:
| FSM states/output coding | Q1 | Q2 |
|---|---|---|
| q0 | 0 | 0 |
| q0 | 0 | 1 |
| q0 | 1 | 0 |
The final mapping from transitions to the biological reporters is then given by:
| J1=0, K1=1, J2=1, K2=0 | J1=0, K1=0, J2=0, K2=1 | J1=1, K1=0, J2=0, K2=1 | J1=0, K1=1, J2=0, K2=1 | J1=0, K1=1, J2=1, K2=0 | J1=0, K1=0, J2=0, K2=1 | J1=0, K1=1, J2=0, K2=0 | J1=0, K1=0, J2=1, K2=1 | J1=1, K1=0, J2=0, K2=0 | J1=1, K1=1, J2=0, K2=0 | J1=1, K1=0, J2=0, K2=1 | J1=0, K1=1, J2=0, K2=1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| cyan | cyan | red | cyan | cyan | cyan | yellow | green | cyan | cyan | cyan | yellow |