Edinburgh/DivisionPopper/Applications
From 2007.igem.org
(→Coupling to a PoPS counting device) |
Luca.Gerosa (Talk | contribs) |
||
| (46 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | '''MENU''' :[[Edinburgh/DivisionPopper| Introduction]] | [[Edinburgh/DivisionPopper/References|Background]] | [[Edinburgh/DivisionPopper/Applications|Applications]] | [[Edinburgh/DivisionPopper/Design|Design&Implementation]] | [[Edinburgh/DivisionPopper/Modelling|Modelling]] | [[Edinburgh/DivisionPopper/Status|Wet Lab]] | [[Edinburgh/DivisionPopper/SBApproach|Synthetic Biology Approach]] | [[Edinburgh/DivisionPopper/Conclusions|Conclusions]] | |
| - | + | The Division PoPper is a device designed to be coupled with other devices in order to offer its functionality for more complex systems. The Division PoPper has no signal inputs, so no upstream devices can be connected. Instead it is a generator of output signal, generating a PoPS pulse each time it senses a cell division. In this sense, the use of a standard signal format such as PoPS is an important characteristic for the compositional power and versatility of the device. In the ongoing work of implementing computational ability in cells, we think a device able to "convert" a core physical behaviour (the division) to an information flow (the PoPS pulse) will be of immense interest. Here we detail some potential uses for the Division PoPper when coupled with other devices. | |
| - | + | ||
| - | + | ||
| - | + | ||
__TOC__ | __TOC__ | ||
| - | + | {| width="20%" align="right" style="text-align:center" | |
| + | |- | ||
| + | |'''System view of the Division Counter''' | ||
| + | |- | ||
| + | |[[Image:Withcounter.png|450px]] The Division PoPper device generates a pulse signal, the Counter device memorizes the number of pulse and activates the Report device if necessary. | ||
| + | |} | ||
| - | ==Division | + | ==Division Counting== |
| - | The output of the Division PoPper | + | One possible application is to count the number of divisions of a cell, by coupling the device to a counter. The simplest configuration is to connect the output of the Division PoPper to the input of the counter. For example, the counter designed by ETH Zurich for the 2005 edition of iGEM is able to receive a PoPS pulse signal and to count how many pulses arrive ([https://2006.igem.org/wiki/index.php/ETH_Zurich_2005#Abstract ETH Zurich counter]). We developed a mathematical model that simulates the behaviour of such a system by integrating our ODE model to the ODE model of the ETH counter (details in the [[Edinburgh/DivisionPopper/Modelling|Modelling]] section). |
| - | ==Division | + | '''Kill-Switch''' |
| + | Why might it be important to count cell divisions? For example to trigger cell death after a certain number of divisions to contain the spread of engineered bacteria when released into the environment to complete a task. | ||
| + | |||
| + | '''Controlled diversity and onset''' | ||
| + | Another possibility is to associate a function at each different division number in order to change cell behaviour over time. For example, where microorganisms manufacture therapeutic proteins, cells could be engineered to begin to express the protein after a certain number of cell divisions. The same holds true for any bioreactor - bacterial behaviour changes with the density of available metabolites. In a closed system (e.g. a bioreactor), conditions could be optimised by only allowing production to start after a special population size is reached. Quorum sensing works in a similar way, but the Division PoPper allows complete flexibility with onset timing whereas a quorum sensing system does not. | ||
| + | |||
| + | |||
| + | |||
| + | {| width="20%" align="right" style="text-align:center" | ||
| + | |- | ||
| + | |'''System view of the Division Frequency Analyzer''' | ||
| + | |- | ||
| + | |[[Image:Withfrequency.png|450px]] The Division PoPper device generates a pulse signal and the Frequency analyzer device calculates the frequency. | ||
| + | |} | ||
| + | |||
| + | ==Division Frequency Analysis== | ||
| - | + | The output of the Division PoPper could be linked to an analyzer of frequency. This can simply be implemented by putting slowly degrading protein downstream. Physically, the slow-degradation protein coding sequence may just follow on from the PoPper. The more frequent the divisions, the more often the PoPS pulse would express the protein and thus increase its concentration. Therefore it's possible to associate the quantity of protein to frequency of division. Since the frequency of division is sometimes related to diseases in humans, this could for example be used to trigger an inhibitory cellular reaction when a cell divides too frequently. | |
| - | |||
| - | == | + | ==Bridging the gap between life and silicon technology== |
| - | + | Computers do not reproduce (yet). The DivisionPoPper is based on a fundamental difference between how green engineering ''actually'' works (on hardware system level) and how synthetic biology wants it to work. It is an overlooked subject, because we are still learning to spot the differences between both worlds. Synthetic biology is the attempt to bridge differences so that we can manipulate biology like we conquered silicon. However, if your central analytical unit suddenly multiplies all its circuits, the system is bound to change and can't be treated 'linearly', depending on what calculations it performs. | |
| - | + | If your laptop suddenly elongated and split in two, you'd be a little bit surprised. Nevertheless this is a character trait of the micro-computers we're dealing with. We believe that we can expect to run into some problems in this strange and exotic world were processors stop working for a little while in order to produce a replica of every single circuit. | |
| - | + | And we think that we can make that sort of situation tangible by presenting a readable output that represents this system change. It allows standardisation of original output data. In that way our device bridges a purely natural phenomenon and Silicon. | |
Latest revision as of 04:16, 27 October 2007
MENU : Introduction | Background | Applications | Design&Implementation | Modelling | Wet Lab | Synthetic Biology Approach | Conclusions
The Division PoPper is a device designed to be coupled with other devices in order to offer its functionality for more complex systems. The Division PoPper has no signal inputs, so no upstream devices can be connected. Instead it is a generator of output signal, generating a PoPS pulse each time it senses a cell division. In this sense, the use of a standard signal format such as PoPS is an important characteristic for the compositional power and versatility of the device. In the ongoing work of implementing computational ability in cells, we think a device able to "convert" a core physical behaviour (the division) to an information flow (the PoPS pulse) will be of immense interest. Here we detail some potential uses for the Division PoPper when coupled with other devices.
Contents |
| System view of the Division Counter |
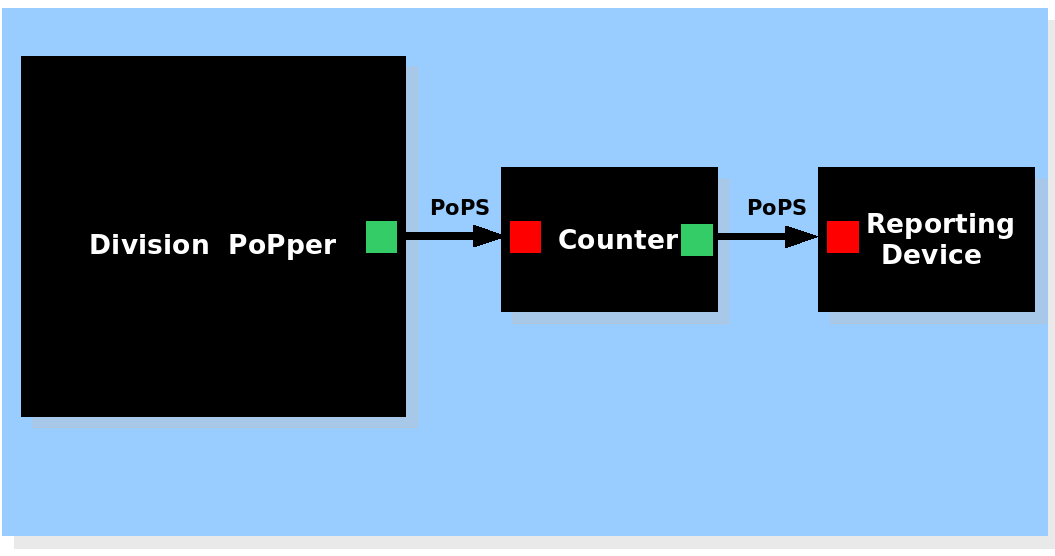 The Division PoPper device generates a pulse signal, the Counter device memorizes the number of pulse and activates the Report device if necessary. The Division PoPper device generates a pulse signal, the Counter device memorizes the number of pulse and activates the Report device if necessary.
|
Division Counting
One possible application is to count the number of divisions of a cell, by coupling the device to a counter. The simplest configuration is to connect the output of the Division PoPper to the input of the counter. For example, the counter designed by ETH Zurich for the 2005 edition of iGEM is able to receive a PoPS pulse signal and to count how many pulses arrive (ETH Zurich counter). We developed a mathematical model that simulates the behaviour of such a system by integrating our ODE model to the ODE model of the ETH counter (details in the Modelling section).
Kill-Switch Why might it be important to count cell divisions? For example to trigger cell death after a certain number of divisions to contain the spread of engineered bacteria when released into the environment to complete a task.
Controlled diversity and onset Another possibility is to associate a function at each different division number in order to change cell behaviour over time. For example, where microorganisms manufacture therapeutic proteins, cells could be engineered to begin to express the protein after a certain number of cell divisions. The same holds true for any bioreactor - bacterial behaviour changes with the density of available metabolites. In a closed system (e.g. a bioreactor), conditions could be optimised by only allowing production to start after a special population size is reached. Quorum sensing works in a similar way, but the Division PoPper allows complete flexibility with onset timing whereas a quorum sensing system does not.
| System view of the Division Frequency Analyzer |
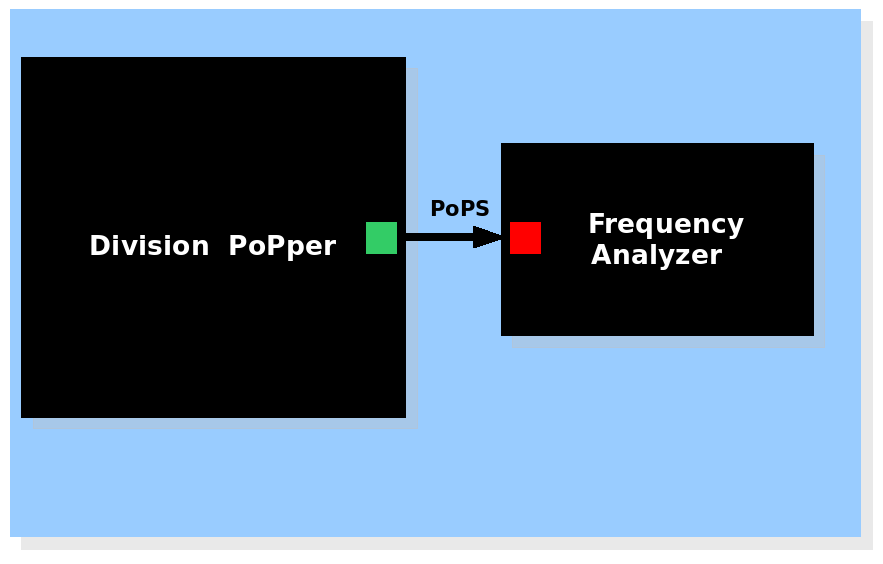 The Division PoPper device generates a pulse signal and the Frequency analyzer device calculates the frequency. The Division PoPper device generates a pulse signal and the Frequency analyzer device calculates the frequency.
|
Division Frequency Analysis
The output of the Division PoPper could be linked to an analyzer of frequency. This can simply be implemented by putting slowly degrading protein downstream. Physically, the slow-degradation protein coding sequence may just follow on from the PoPper. The more frequent the divisions, the more often the PoPS pulse would express the protein and thus increase its concentration. Therefore it's possible to associate the quantity of protein to frequency of division. Since the frequency of division is sometimes related to diseases in humans, this could for example be used to trigger an inhibitory cellular reaction when a cell divides too frequently.
Bridging the gap between life and silicon technology
Computers do not reproduce (yet). The DivisionPoPper is based on a fundamental difference between how green engineering actually works (on hardware system level) and how synthetic biology wants it to work. It is an overlooked subject, because we are still learning to spot the differences between both worlds. Synthetic biology is the attempt to bridge differences so that we can manipulate biology like we conquered silicon. However, if your central analytical unit suddenly multiplies all its circuits, the system is bound to change and can't be treated 'linearly', depending on what calculations it performs.
If your laptop suddenly elongated and split in two, you'd be a little bit surprised. Nevertheless this is a character trait of the micro-computers we're dealing with. We believe that we can expect to run into some problems in this strange and exotic world were processors stop working for a little while in order to produce a replica of every single circuit.
And we think that we can make that sort of situation tangible by presenting a readable output that represents this system change. It allows standardisation of original output data. In that way our device bridges a purely natural phenomenon and Silicon.